Aberrant T cell differentiation in the absence of Dicer
- PMID: 16009718
- PMCID: PMC2212998
- DOI: 10.1084/jem.20050678
Aberrant T cell differentiation in the absence of Dicer
Abstract
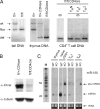
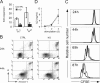
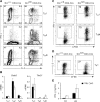
Dicer is an RNaseIII-like enzyme that is required for generating short interfering RNAs and microRNAs. The latter have been implicated in regulating cell fate determination in invertebrates and vertebrates. To test the requirement for Dicer in cell-lineage decisions in a mammalian organism, we have generated a conditional allele of dicer-1 (dcr-1) in the mouse. Specific deletion of dcr-1 in the T cell lineage resulted in impaired T cell development and aberrant T helper cell differentiation and cytokine production. A severe block in peripheral CD8(+) T cell development was observed upon dcr-1 deletion in the thymus. However, Dicer-deficient CD4(+) T cells, although reduced in numbers, were viable and could be analyzed further. These cells were defective in microRNA processing, and upon stimulation they proliferated poorly and underwent increased apoptosis. Independent of their proliferation defect, Dicer-deficient helper T cells preferentially expressed interferon-gamma, the hallmark effector cytokine of the Th1 lineage.
Figures



Similar articles
-
T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer.J Exp Med. 2005 May 2;201(9):1367-73. doi: 10.1084/jem.20050572. J Exp Med. 2005. PMID: 15867090 Free PMC article.
-
Constitutive expression of CIITA directs CD4 T cells to produce Th2 cytokines in the thymus.Cell Immunol. 2005 Jan;233(1):30-40. doi: 10.1016/j.cellimm.2005.03.006. Cell Immunol. 2005. PMID: 15876426
-
Dicer-dependent microRNA pathway controls invariant NKT cell development.J Immunol. 2009 Aug 15;183(4):2506-12. doi: 10.4049/jimmunol.0901361. Epub 2009 Jul 22. J Immunol. 2009. PMID: 19625646
-
Dicer Regulates the Balance of Short-Lived Effector and Long-Lived Memory CD8 T Cell Lineages.PLoS One. 2016 Sep 14;11(9):e0162674. doi: 10.1371/journal.pone.0162674. eCollection 2016. PLoS One. 2016. PMID: 27627450 Free PMC article.
-
MicroRNAs are key regulators controlling iNKT and regulatory T-cell development and function.Cell Mol Immunol. 2011 Sep;8(5):380-7. doi: 10.1038/cmi.2011.27. Epub 2011 Aug 8. Cell Mol Immunol. 2011. PMID: 21822298 Free PMC article. Review.
Cited by
-
Plasticity within the αβ⁺CD4⁺ T-cell lineage: when, how and what for?Open Biol. 2013 Jan 23;3(1):120157. doi: 10.1098/rsob.120157. Open Biol. 2013. PMID: 23345540 Free PMC article. Review.
-
An evolutionarily conserved mutual interdependence between Aire and microRNAs in promiscuous gene expression.Eur J Immunol. 2013 Jul;43(7):1769-78. doi: 10.1002/eji.201343343. Epub 2013 May 17. Eur J Immunol. 2013. PMID: 23589212 Free PMC article.
-
Regulation of lipid metabolism by Dicer revealed through SILAC mice.J Proteome Res. 2012 Apr 6;11(4):2193-205. doi: 10.1021/pr2009884. Epub 2012 Feb 28. J Proteome Res. 2012. PMID: 22313051 Free PMC article.
-
T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire.J Exp Med. 2013 Feb 11;210(2):417-32. doi: 10.1084/jem.20111717. Epub 2013 Feb 4. J Exp Med. 2013. PMID: 23382546 Free PMC article.
-
Small RNAs and RNAi pathways in meiotic prophase I.Chromosome Res. 2007;15(5):653-65. doi: 10.1007/s10577-007-1144-z. Chromosome Res. 2007. PMID: 17674152 Review.
References
-
- Mello, C.C., and D. Conte Jr. 2004. Revealing the world of RNA interference. Nature. 431:338–342. - PubMed
-
- Ambros, V. 2004. The functions of animal microRNAs. Nature. 431:350–355. - PubMed
-
- Lippman, Z., and R. Martienssen. 2004. The role of RNA interference in heterochromatic silencing. Nature. 431:364–370. - PubMed
-
- Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature. 431:343–349. - PubMed
-
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

