The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element
- PMID: 15987813
- PMCID: PMC1370787
- DOI: 10.1261/rna.7218905
The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element
Abstract
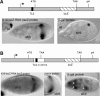
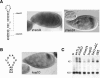
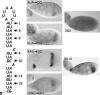
The subcellular localization of mRNAs is a key step in the polarization of cells in organisms from yeast to man. Here, we use a transgenic fly/in situ hybridization assay system to define the positional, structural, and sequence requirements of the TLS, a stem loop RNA sequence element that mediates the subcellular localization of K10 and Orb transcripts in Drosophila oocytes. We find that the TLS is a highly robust and modular element. It mediates efficient RNA localization regardless of sequence context or position within the transcript. Site-specific mutagenesis experiments indicate that the size and shape of the stem and loop regions are critical determinants of TLS activity. Such experiments also identify specific base residues that are important for TLS activity. All such residues map to the stem portion of the structure. Significantly, mutations at these residues interfere with TLS activity only when they alter the stereochemistry of the stem's minor groove. For example, mutation of the A:U base pair at position 3 of the TLS stem to G:C severely reduces TLS activity, while mutation of the same base pair to U:A has no effect. Extensive searches for TLS-like elements in other Drosophila mRNAs using sequence and structural parameters defined by our experiments indicate that the TLS is unique to K10 and Orb mRNAs. This unexpected finding raises important questions as to how the many hundreds of other mRNAs that are known or thought to exhibit K10 and Orb-like localization are localized.
Figures




Similar articles
-
A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA.Development. 1995 Nov;121(11):3809-18. doi: 10.1242/dev.121.11.3809. Development. 1995. PMID: 8582290
-
Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II.Mol Cell Biol. 1995 Aug;15(8):4562-71. doi: 10.1128/MCB.15.8.4562. Mol Cell Biol. 1995. PMID: 7623847 Free PMC article.
-
Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression.Genes Dev. 2001 Jan 15;15(2):173-87. doi: 10.1101/gad.862801. Genes Dev. 2001. PMID: 11157774 Free PMC article.
-
The Translin/Trax RNA binding complex: clues to function in the nervous system.Biochim Biophys Acta. 2008 Aug;1779(8):479-85. doi: 10.1016/j.bbagrm.2008.03.008. Epub 2008 Apr 3. Biochim Biophys Acta. 2008. PMID: 18424275 Free PMC article. Review.
-
A glimpse of the machinery.Curr Biol. 1999 Oct 7;9(19):R725-7. doi: 10.1016/s0960-9822(99)80468-0. Curr Biol. 1999. PMID: 10530994 Review.
Cited by
-
A'-form RNA helices are required for cytoplasmic mRNA transport in Drosophila.Nat Struct Mol Biol. 2010 Jun;17(6):703-9. doi: 10.1038/nsmb.1813. Epub 2010 May 16. Nat Struct Mol Biol. 2010. PMID: 20473315 Free PMC article.
-
Evidence for a transport-trap mode of Drosophila melanogaster gurken mRNA localization.PLoS One. 2010 Nov 12;5(11):e15448. doi: 10.1371/journal.pone.0015448. PLoS One. 2010. PMID: 21103393 Free PMC article.
-
Translational co-regulation of a ligand and inhibitor by a conserved RNA element.Nucleic Acids Res. 2018 Jan 9;46(1):104-119. doi: 10.1093/nar/gkx938. Nucleic Acids Res. 2018. PMID: 29059375 Free PMC article.
-
Identification of 3'UTR sequence elements and a teloplasm localization motif sufficient for the localization of Hro-twist mRNA to the zygotic animal and vegetal poles.Dev Growth Differ. 2012 May;54(4):519-34. doi: 10.1111/j.1440-169X.2012.01352.x. Dev Growth Differ. 2012. PMID: 22587329 Free PMC article.
-
Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles.BMC Genomics. 2011 Nov 30;12 Suppl 3(Suppl 3):S18. doi: 10.1186/1471-2164-12-S3-S18. Epub 2011 Nov 30. BMC Genomics. 2011. PMID: 22369587 Free PMC article.
References
-
- Arn, E.A., Cha, B.J., Theurkauf, W.E., and Macdonald, P.M. 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4: 41–51. - PubMed
-
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 1998. RNA localization in development. Annu. Rev. Biochem. 67: 335–394. - PubMed
-
- Bullock, S.L. and Ish-Horowicz, D. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414: 611–616. - PubMed
-
- Cheung, H.-K., Serano, T.L., and Cohen, R.S. 1992. Evidence for a highly selective RNA transport system and its role in establishing the dorsoventral axis of the Drosophila egg. Development 114: 653–661. - PubMed
-
- Cohen, R.S. and Meselson, M. 1985. Separate regulatory elements for the heat-inducible and ovarian expression of the Drosophila hsp26 gene. Cell 42: 737–746. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
