GlyProt: in silico glycosylation of proteins
- PMID: 15980456
- PMCID: PMC1160146
- DOI: 10.1093/nar/gki385
GlyProt: in silico glycosylation of proteins
Abstract
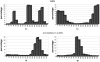
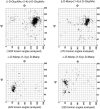
GlyProt (http://www.glycosciences.de/glyprot/) is a web-based tool that enables meaningful N-glycan conformations to be attached to all the spatially accessible potential N-glycosylation sites of a known three-dimensional (3D) protein structure. The probabilities of physicochemical properties such as mass, accessible surface and radius of gyration are calculated. The purpose of this service is to provide rapid access to reliable 3D models of glycoproteins, which can subsequently be refined by using more elaborate simulations and validated by comparing the generated models with experimental data.
Figures



Similar articles
-
Glycosylator: a Python framework for the rapid modeling of glycans.BMC Bioinformatics. 2019 Oct 22;20(1):513. doi: 10.1186/s12859-019-3097-6. BMC Bioinformatics. 2019. PMID: 31640540 Free PMC article.
-
Glycosylation of proteins: a computer based method for the rapid exploration of conformational space of N-glycans.Pac Symp Biocomput. 2002:285-96. doi: 10.1142/9789812799623_0027. Pac Symp Biocomput. 2002. PMID: 11928483
-
GlycoPP: a webserver for prediction of N- and O-glycosites in prokaryotic protein sequences.PLoS One. 2012;7(7):e40155. doi: 10.1371/journal.pone.0040155. Epub 2012 Jul 9. PLoS One. 2012. PMID: 22808107 Free PMC article.
-
GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research.Glycobiology. 2006 May;16(5):71R-81R. doi: 10.1093/glycob/cwj049. Epub 2005 Oct 20. Glycobiology. 2006. PMID: 16239495 Review.
-
Methods in enzymology: O-glycosylation of proteins.Methods Enzymol. 2005;405:139-71. doi: 10.1016/S0076-6879(05)05007-X. Methods Enzymol. 2005. PMID: 16413314 Review.
Cited by
-
Using databases and web resources for glycomics research.Mol Cell Proteomics. 2013 Apr;12(4):1036-45. doi: 10.1074/mcp.R112.026252. Epub 2013 Jan 16. Mol Cell Proteomics. 2013. PMID: 23325765 Free PMC article. Review.
-
Crystal structure of the conserved domain of the DC lysosomal associated membrane protein: implications for the lysosomal glycocalyx.BMC Biol. 2012 Jul 19;10:62. doi: 10.1186/1741-7007-10-62. BMC Biol. 2012. PMID: 22809326 Free PMC article.
-
Glycans on influenza hemagglutinin affect receptor binding and immune response.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18137-42. doi: 10.1073/pnas.0909696106. Epub 2009 Oct 12. Proc Natl Acad Sci U S A. 2009. PMID: 19822741 Free PMC article.
-
Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein.Acta Crystallogr D Biol Crystallogr. 2009 Nov;65(Pt 11):1162-80. doi: 10.1107/S0907444909032314. Epub 2009 Oct 22. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19923712 Free PMC article.
-
Polymorphisms and interspecies differences of the activating and inhibitory FcγRII of Macaca nemestrina influence the binding of human IgG subclasses.J Immunol. 2014 Jan 15;192(2):792-803. doi: 10.4049/jimmunol.1301554. Epub 2013 Dec 16. J Immunol. 2014. PMID: 24342805 Free PMC article.
References
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. - PubMed
-
- Ben-Dor S., Esterman N., Rubin E. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. - PubMed
-
- Luetteke T., Frank M., von der Lieth C.W. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res. 2004;339:1015–1020. - PubMed
-
- Imberty A., Perez S. Structure, conformation, and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem. Rev. 2000;100:4567–4588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

