A molecular docking model of SARS-CoV S1 protein in complex with its receptor, human ACE2
- PMID: 15979045
- PMCID: PMC7106554
- DOI: 10.1016/j.compbiolchem.2005.04.008
A molecular docking model of SARS-CoV S1 protein in complex with its receptor, human ACE2
Abstract
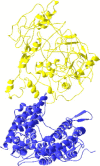
The exact residues within severe acute respiratory syndrome coronavirus (SARS-CoV) S1 protein and its receptor, human ACE2, involved in their interaction still remain largely undetermined. Identification of exact amino acid residues that are crucial for the interaction of S1 with ACE2 could provide working hypotheses for experimental studies and might be helpful for the development of antiviral inhibitor. In this paper, a molecular docking model of SARS-CoV S1 protein in complex with human ACE2 was constructed. The interacting residue pairs within this complex model and their contact types were also identified. Our model, supported by significant biochemical evidence, suggested receptor-binding residues were concentrated in two segments of S1 protein. In contrast, the interfacial residues in ACE2, though close to each other in tertiary structure, were found to be widely scattered in the primary sequence. In particular, the S1 residue ARG453 and ACE2 residue LYS341 might be the key residues in the complex formation.
Figures
Similar articles
-
A model of the ACE2 structure and function as a SARS-CoV receptor.Biochem Biophys Res Commun. 2004 Jan 30;314(1):235-41. doi: 10.1016/j.bbrc.2003.12.081. Biochem Biophys Res Commun. 2004. PMID: 14715271 Free PMC article.
-
Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2.PLoS Pathog. 2018 Aug 13;14(8):e1007236. doi: 10.1371/journal.ppat.1007236. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30102747 Free PMC article.
-
Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.Science. 2005 Sep 16;309(5742):1864-8. doi: 10.1126/science.1116480. Science. 2005. PMID: 16166518
-
Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus.Cell Mol Life Sci. 2004 Nov;61(21):2738-43. doi: 10.1007/s00018-004-4242-5. Cell Mol Life Sci. 2004. PMID: 15549175 Free PMC article. Review.
-
Biological, clinical and epidemiological features of COVID-19, SARS and MERS and AutoDock simulation of ACE2.Infect Dis Poverty. 2020 Jul 20;9(1):99. doi: 10.1186/s40249-020-00691-6. Infect Dis Poverty. 2020. PMID: 32690096 Free PMC article. Review.
Cited by
-
Targeting Viral ORF3a Protein: A New Approach to Mitigate COVID-19 Induced Immune Cell Apoptosis and Associated Respiratory Complications.Adv Pharm Bull. 2023 Nov;13(4):678-687. doi: 10.34172/apb.2023.069. Epub 2023 Jan 23. Adv Pharm Bull. 2023. PMID: 38022818 Free PMC article. Review.
-
Current Status of Microneedle Array Technology for Therapeutic Delivery: From Bench to Clinic.Mol Biotechnol. 2024 Dec;66(12):3415-3437. doi: 10.1007/s12033-023-00961-2. Epub 2023 Nov 21. Mol Biotechnol. 2024. PMID: 37987985 Review.
-
Drug Repurposing Against Angiotensin-Converting Enzyme-Related Carboxypeptidase (ACE2) Through Computational Approach.J Med Signals Sens. 2022 Nov 10;12(4):341-346. doi: 10.4103/jmss.JMSS_66_20. eCollection 2022 Oct-Dec. J Med Signals Sens. 2022. PMID: 36726422 Free PMC article.
-
Mechanisms of SARS-CoV-2 Infection-Induced Kidney Injury: A Literature Review.Front Cell Infect Microbiol. 2022 Jun 14;12:838213. doi: 10.3389/fcimb.2022.838213. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35774397 Free PMC article. Review.
-
The effect of various compounds on the COVID mechanisms, from chemical to molecular aspects.Biophys Chem. 2022 Sep;288:106824. doi: 10.1016/j.bpc.2022.106824. Epub 2022 May 12. Biophys Chem. 2022. PMID: 35728510 Free PMC article. Review.
References
-
- Cai Q.C., Jiang Q.W., Zhao G.M., Guo Q., Cao G.W., Chen T. Putative caveolin-binding sites in SARS-CoV proteins. Acta Pharmacol. Sin. 2003;24:1051–1059. - PubMed
-
- Chen R., Li L., Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. - PubMed
-
- Chen R., Tong W., Mintseris J., Li L., Weng Z. ZDOCK predictions for the CAPRI challenge. Proteins. 2003;52:68–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

