Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice
- PMID: 15972459
- PMCID: PMC1456781
- DOI: 10.1534/genetics.105.043612
Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice
Abstract
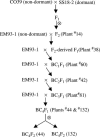
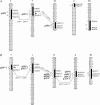
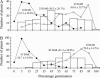
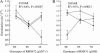
Association of seed dormancy with shattering, awn, and black hull and red pericarp colors enhances survival of wild and weedy species, but challenges the use of dormancy genes in breeding varieties resistant to preharvest sprouting. A phenotypic selection and recurrent backcrossing technique was used to introduce dormancy genes from a wild-like weedy rice to a breeding line to determine their effects and linkage with the other traits. Five generations of phenotypic selection alone for low germination extremes simultaneously retained dormancy alleles at five independent QTL, including qSD12 (R(2) > 50%), as determined by genome-wide scanning for their main and/or epistatic effects in two BC(4)F(2) populations. Four dormancy loci with moderate to small effects colocated with QTL/genes for one to three of the associated traits. Multilocus response to the selection suggests that these dormancy genes are cumulative in effect, as well as networked by epistases, and that the network may have played a "sheltering" role in maintaining intact adaptive haplotypes during the evolution of weeds. Tight linkage may prevent the dormancy genes from being used in breeding programs. The major effect of qSD12 makes it an ideal target for map-based cloning and the best candidate for imparting resistance to preharvest sprouting.
Figures

Similar articles
-
Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa).Theor Appl Genet. 2005 Apr;110(6):1108-18. doi: 10.1007/s00122-005-1939-2. Epub 2005 Mar 22. Theor Appl Genet. 2005. PMID: 15782297
-
Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa).Genetics. 2004 Mar;166(3):1503-16. doi: 10.1534/genetics.166.3.1503. Genetics. 2004. PMID: 15082564 Free PMC article.
-
Comparative Mapping of Seed Dormancy Loci Between Tropical and Temperate Ecotypes of Weedy Rice (Oryza sativa L.).G3 (Bethesda). 2017 Aug 7;7(8):2605-2614. doi: 10.1534/g3.117.040451. G3 (Bethesda). 2017. PMID: 28592557 Free PMC article.
-
[Major domestication traits in Asian rice].Yi Chuan. 2012 Nov;34(11):1379-89. doi: 10.3724/sp.j.1005.2012.01379. Yi Chuan. 2012. PMID: 23208135 Review. Chinese.
-
Advances in Rice Seed Shattering.Int J Mol Sci. 2023 May 17;24(10):8889. doi: 10.3390/ijms24108889. Int J Mol Sci. 2023. PMID: 37240235 Free PMC article. Review.
Cited by
-
Quantitative trait locus and haplotype analyses of wild and crop-mimic traits in U.S. weedy rice.G3 (Bethesda). 2013 Jun 21;3(6):1049-59. doi: 10.1534/g3.113.006395. G3 (Bethesda). 2013. PMID: 23604075 Free PMC article.
-
Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication.Plant Cell. 2008 Nov;20(11):2946-59. doi: 10.1105/tpc.108.060426. Epub 2008 Nov 25. Plant Cell. 2008. PMID: 19033526 Free PMC article.
-
Multiple origins of the phenol reaction negative phenotype in foxtail millet, Setaria italica (L.) P. Beauv., were caused by independent loss-of-function mutations of the polyphenol oxidase (Si7PPO) gene during domestication.Mol Genet Genomics. 2015 Aug;290(4):1563-74. doi: 10.1007/s00438-015-1022-x. Epub 2015 Mar 5. Mol Genet Genomics. 2015. PMID: 25740049
-
An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice.Plant Cell. 2013 Sep;25(9):3360-76. doi: 10.1105/tpc.113.113589. Epub 2013 Sep 27. Plant Cell. 2013. PMID: 24076974 Free PMC article.
-
Dormancy genes from weedy rice respond divergently to seed development environments.Genetics. 2006 Feb;172(2):1199-211. doi: 10.1534/genetics.105.049155. Epub 2005 Nov 4. Genetics. 2006. PMID: 16272412 Free PMC article.
References
-
- Ahn, S. N., J. P. Suh, C. S. Oh, S. J. Lee and H. S. Suh, 2002. Development of introgression lines of weedy rice in the background of Tongil-type rice. Rice Genet. Newsl. 19: 14–15.
-
- Anderson, J. A., M. E. Sorrells and S. D. Tanksley, 1993. RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci. 33: 453–459.
-
- Baker, B., D. V. Chin and M. Mortimer, 2000. Wild and Weedy Rice in Rice Ecosystems in Asia: A Review. Limited Proceedings 2000, No. 2, International Rice Research Institute, Manila, Philippines.
-
- Basu, C., M. D. Halfhill, T. C. Mueller and C. N. Jr. Stewart, 2004. Weed genomics: new tools to understand weed biology. Trends Plant Sci. 9: 391–398. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

