Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50)
- PMID: 15956592
- PMCID: PMC1143749
- DOI: 10.1128/JVI.79.13.8493-8505.2005
Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50)
Abstract
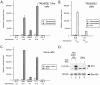
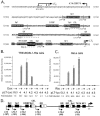
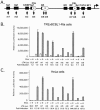
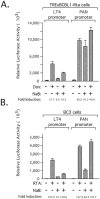
Kaposi's sarcoma-associated herpesvirus (KSHV) maintains a latent infection in primary effusion lymphoma cells but can be induced to enter full lytic replication by exposure to a variety of chemical inducing agents or by expression of the KSHV-encoded replication and transcription activator (RTA) protein. During latency, only a few viral genes are expressed, and these include the three genes of the so-called latency transcript (LT) cluster: v-FLIP (open reading frame 71 [ORF71]), v-cyclin (ORF72), and latency-associated nuclear antigen (ORF73). During latency, all three open reading frames are transcribed from a common promoter as part of a multicistronic mRNA. Subsequent alternative mRNA splicing and internal ribosome entry allows for the expression of each protein. Here, we show that transcription of LT cassette mRNA can be induced by RTA through the activation of a second promoter (LT(i)) immediately downstream of the constitutively active promoter (LT(c)). We identified a minimal cis-regulatory region, which overlaps with the promoter for the bicistronic K14/v-GPCR delayed early gene that is transcribed in the opposite direction. In addition to a TATA box at -30 relative to the LT(i) mRNA start sites, we identified three separate RTA response elements that are also utilized by the K14/v-GPCR promoter. Interestingly, LT(i) is unresponsive to sodium butyrate, a potent inducer of lytic replication. This suggests there is a previously unrecognized class of RTA-responsive promoters that respond to direct, but not indirect, induction of RTA. These studies highlight the fact that induction method can influence the precise program of viral gene expression during early events in reactivation and also suggest a mechanism by which RTA contributes to establishment of latency during de novo infections.
Figures
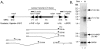


Similar articles
-
CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters.J Virol. 2003 Sep;77(17):9590-612. doi: 10.1128/jvi.77.17.9590-9612.2003. J Virol. 2003. PMID: 12915572 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency.J Virol. 2004 Jun;78(12):6585-94. doi: 10.1128/JVI.78.12.6585-6594.2004. J Virol. 2004. PMID: 15163750 Free PMC article.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
Cited by
-
ARID3B: a Novel Regulator of the Kaposi's Sarcoma-Associated Herpesvirus Lytic Cycle.J Virol. 2016 Sep 29;90(20):9543-55. doi: 10.1128/JVI.03262-15. Print 2016 Oct 15. J Virol. 2016. PMID: 27512077 Free PMC article.
-
Cyclooxygenase 2 induced by Kaposi's sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression.J Virol. 2006 Jul;80(13):6534-52. doi: 10.1128/JVI.00231-06. J Virol. 2006. PMID: 16775340 Free PMC article.
-
Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators.Oncotarget. 2017 Sep 5;8(53):91425-91444. doi: 10.18632/oncotarget.20648. eCollection 2017 Oct 31. Oncotarget. 2017. PMID: 29207655 Free PMC article.
-
Wide-scale use of Notch signaling factor CSL/RBP-Jkappa in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes.J Virol. 2010 Feb;84(3):1334-47. doi: 10.1128/JVI.01301-09. Epub 2009 Nov 11. J Virol. 2010. PMID: 19906914 Free PMC article.
-
KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features.PLoS Pathog. 2014 Jan;10(1):e1003847. doi: 10.1371/journal.ppat.1003847. Epub 2014 Jan 16. PLoS Pathog. 2014. PMID: 24453964 Free PMC article.
References
-
- Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. - PubMed
-
- AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. - PubMed
-
- Byun, H., Y. Gwack, S. Hwang, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame (ORF) 50 transactivates K8 and ORF57 promoters via heterogeneous response elements. Mol. Cells 14:185-191. - PubMed
-
- Cesarman, E. 2003. Kaposi's sarcoma-associated herpesvirus-the high cost of viral survival. N. Engl. J. Med. 349:1107-1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

