Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly
- PMID: 15956550
- PMCID: PMC1143739
- DOI: 10.1128/JVI.79.13.8046-8056.2005
Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly
Abstract
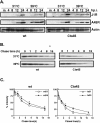
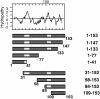
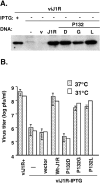
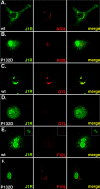
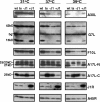
Vaccinia virus J1R protein is required for virion morphogenesis (W. L. Chiu and W. Chang, J. Virol. 76:9575-9587, 2002). In this work, we further characterized the J1R protein of wild-type vaccinia virus and compared it with the protein encoded by the temperature-sensitive mutant virus Cts45. The mutant Cts45 was found to contain a Pro-to-Ser substitution at residue 132 of the J1R open reading frame, which is responsible for a loss-of-function phenotype. The half-life of the J1R-P132S mutant protein was comparable at both 31 and 39 degrees C, indicating that the P132S mutation did not affect the stability of the J1R protein. We also showed that the J1R protein interacts with itself in the virus-infected cells. The N-terminal region of the J1R protein, amino acids (aa) 1 to 77, interacted with the C-terminal region, aa 84 to 153, and the P132 mutation did not abolish this interaction, as determined by two-hybrid analysis. Furthermore, we demonstrated that J1R protein is part of a viral complex containing the A30L, G7L, and F10L proteins in virus-infected cells. In immunofluorescence analyses, wild-type J1R protein colocalized with the A30L, G7L, and F10L proteins in virus-infected cells but the loss-of-function P132 mutant did not. Furthermore, without a functional J1R protein, rapid degradation of A30L and the 15-kDa forms of the G7L and F10L proteins was observed in cells infected with Cts45 at 39 degrees C. This study thus demonstrated the importance of the J1R protein in the formation of a viral assembly complex required for morphogenesis.
Figures


Similar articles
-
Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis.J Virol. 2002 Oct;76(19):9575-87. doi: 10.1128/jvi.76.19.9575-9587.2002. J Virol. 2002. PMID: 12208937 Free PMC article.
-
Unique temperature-sensitive defect in vaccinia virus morphogenesis maps to a single nucleotide substitution in the A30L gene.J Virol. 2001 Nov;75(22):11222-6. doi: 10.1128/JVI.75.22.11222-11226.2001. J Virol. 2001. PMID: 11602762 Free PMC article.
-
Vaccinia virus G7L protein Interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions.J Virol. 2003 Mar;77(6):3418-29. doi: 10.1128/jvi.77.6.3418-3429.2003. J Virol. 2003. PMID: 12610117 Free PMC article.
-
Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature.J Virol. 2004 Oct;78(19):10238-48. doi: 10.1128/JVI.78.19.10238-10248.2004. J Virol. 2004. PMID: 15367589 Free PMC article.
-
Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants.J Virol. 2001 Feb;75(4):1656-63. doi: 10.1128/JVI.75.4.1656-1663.2001. J Virol. 2001. PMID: 11160663 Free PMC article.
Cited by
-
Expression of the highly conserved vaccinia virus E6 protein is required for virion morphogenesis.Virology. 2009 Apr 10;386(2):478-85. doi: 10.1016/j.virol.2009.01.009. Epub 2009 Feb 12. Virology. 2009. PMID: 19217136 Free PMC article.
-
Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants.Virology. 2008 May 25;375(1):213-22. doi: 10.1016/j.virol.2008.01.027. Epub 2008 Mar 7. Virology. 2008. PMID: 18314155 Free PMC article.
-
A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication.Adv Virus Res. 2007;70:101-82. doi: 10.1016/S0065-3527(07)70004-0. Adv Virus Res. 2007. PMID: 17765705 Free PMC article. Review.
-
From crescent to mature virion: vaccinia virus assembly and maturation.Viruses. 2014 Oct 7;6(10):3787-808. doi: 10.3390/v6103787. Viruses. 2014. PMID: 25296112 Free PMC article. Review.
-
Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles.J Virol. 2006 Mar;80(5):2127-40. doi: 10.1128/JVI.80.5.2127-2140.2006. J Virol. 2006. PMID: 16474121 Free PMC article.
References
-
- Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241-263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

