Altered nucleosome occupancy and histone H3K4 methylation in response to 'transcriptional stress'
- PMID: 15944735
- PMCID: PMC1173152
- DOI: 10.1038/sj.emboj.7600711
Altered nucleosome occupancy and histone H3K4 methylation in response to 'transcriptional stress'
Abstract
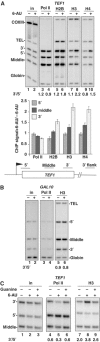
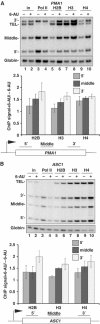
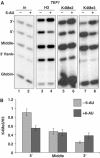
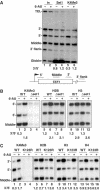
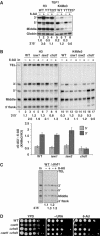
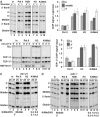
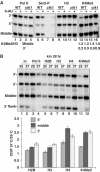
We report that under 'transcriptional stress' in budding yeast, when most pol II activity is acutely inhibited, rapid deposition of nucleosomes occurs within genes, particularly at 3' positions. Whereas histone H3K4 trimethylation normally marks 5' ends of highly transcribed genes, under 'transcriptional stress' induced by 6-azauracil (6-AU) and inactivation of pol II, TFIIE or CTD kinases Kin28 and Ctk1, this mark shifted to the 3' end of the TEF1 gene. H3K4Me3 at 3' positions was dynamic and could be rapidly removed when transcription recovered. Set1 and Chd1 are required for H3K4 trimethylation at 3' positions when transcription is inhibited by 6-AU. Furthermore, Deltachd1 suppressed the growth defect of Deltaset1. We suggest that a 'transcriptional stress' signal sensed through Set1, Chd1, and possibly other factors, causes H3K4 hypermethylation of newly deposited nucleosomes at downstream positions within a gene. This response identifies a new role for H3K4 trimethylation at the 3' end of the gene, as a chromatin mark associated with impaired pol II transcription.
Figures

Similar articles
-
The RNA polymerase II kinase Ctk1 regulates positioning of a 5' histone methylation boundary along genes.Mol Cell Biol. 2007 Jan;27(2):721-31. doi: 10.1128/MCB.01628-06. Epub 2006 Nov 6. Mol Cell Biol. 2007. PMID: 17088384 Free PMC article.
-
Ctk complex-mediated regulation of histone methylation by COMPASS.Mol Cell Biol. 2007 Jan;27(2):709-20. doi: 10.1128/MCB.01627-06. Epub 2006 Nov 6. Mol Cell Biol. 2007. PMID: 17088385 Free PMC article.
-
H3K4 Methylation Dependent and Independent Chromatin Regulation by JHD2 and SET1 in Budding Yeast.G3 (Bethesda). 2018 May 4;8(5):1829-1839. doi: 10.1534/g3.118.200151. G3 (Bethesda). 2018. PMID: 29599176 Free PMC article.
-
The multiple faces of Set1.Biochem Cell Biol. 2006 Aug;84(4):536-48. doi: 10.1139/o06-081. Biochem Cell Biol. 2006. PMID: 16936826 Review.
-
Can't remember to forget you: Chromatin-based priming of somatic stress responses.Semin Cell Dev Biol. 2018 Nov;83:133-139. doi: 10.1016/j.semcdb.2017.09.032. Epub 2017 Sep 29. Semin Cell Dev Biol. 2018. PMID: 28951121 Review.
Cited by
-
The RNA polymerase II kinase Ctk1 regulates positioning of a 5' histone methylation boundary along genes.Mol Cell Biol. 2007 Jan;27(2):721-31. doi: 10.1128/MCB.01628-06. Epub 2006 Nov 6. Mol Cell Biol. 2007. PMID: 17088384 Free PMC article.
-
TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II.Mol Cell Biol. 2009 Oct;29(20):5455-64. doi: 10.1128/MCB.00637-09. Epub 2009 Aug 10. Mol Cell Biol. 2009. PMID: 19667075 Free PMC article.
-
Ready, SET, Go: Post-translational regulation of the histone lysine methylation network in budding yeast.J Biol Chem. 2021 Aug;297(2):100939. doi: 10.1016/j.jbc.2021.100939. Epub 2021 Jul 3. J Biol Chem. 2021. PMID: 34224729 Free PMC article. Review.
-
Functional role of histone variant Htz1 in the stress response to oleate in Saccharomyces cerevisiae.Biosci Rep. 2015 May 20;35(4):e00224. doi: 10.1042/BSR20150114. Biosci Rep. 2015. PMID: 26182431 Free PMC article.
-
Multi-tasking on chromatin with the SAGA coactivator complexes.Mutat Res. 2007 May 1;618(1-2):135-48. doi: 10.1016/j.mrfmmm.2006.09.008. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17337012 Free PMC article. Review.
References
-
- Adkins MW, Howar SR, Tyler JK (2004) Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 - PubMed
-
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 - PubMed
-
- Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 10: 1441–1452 - PubMed
-
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

