The TetR family of transcriptional repressors
- PMID: 15944459
- PMCID: PMC1197418
- DOI: 10.1128/MMBR.69.2.326-356.2005
The TetR family of transcriptional repressors
Abstract
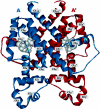
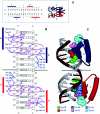
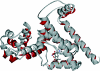
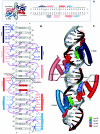
We have developed a general profile for the proteins of the TetR family of repressors. The stretch that best defines the profile of this family is made up of 47 amino acid residues that correspond to the helix-turn-helix DNA binding motif and adjacent regions in the three-dimensional structures of TetR, QacR, CprB, and EthR, four family members for which the function and three-dimensional structure are known. We have detected a set of 2,353 nonredundant proteins belonging to this family by screening genome and protein databases with the TetR profile. Proteins of the TetR family have been found in 115 genera of gram-positive, alpha-, beta-, and gamma-proteobacteria, cyanobacteria, and archaea. The set of genes they regulate is known for 85 out of the 2,353 members of the family. These proteins are involved in the transcriptional control of multidrug efflux pumps, pathways for the biosynthesis of antibiotics, response to osmotic stress and toxic chemicals, control of catabolic pathways, differentiation processes, and pathogenicity. The regulatory network in which the family member is involved can be simple, as in TetR (i.e., TetR bound to the target operator represses tetA transcription and is released in the presence of tetracycline), or more complex, involving a series of regulatory cascades in which either the expression of the TetR family member is modulated by another regulator or the TetR family member triggers a cell response to react to environmental insults. Based on what has been learned from the cocrystals of TetR and QacR with their target operators and from their three-dimensional structures in the absence and in the presence of ligands, and based on multialignment analyses of the conserved stretch of 47 amino acids in the 2,353 TetR family members, two groups of residues have been identified. One group includes highly conserved positions involved in the proper orientation of the helix-turn-helix motif and hence seems to play a structural role. The other set of less conserved residues are involved in establishing contacts with the phosphate backbone and target bases in the operator. Information related to the TetR family of regulators has been updated in a database that can be accessed at www.bactregulators.org.
Figures




Similar articles
-
Mechanisms underlying expression of Tn10 encoded tetracycline resistance.Annu Rev Microbiol. 1994;48:345-69. doi: 10.1146/annurev.mi.48.100194.002021. Annu Rev Microbiol. 1994. PMID: 7826010 Review.
-
Combinations of the alpha-helix-turn-alpha-helix motif of TetR with respective residues from LacI or 434Cro: DNA recognition, inducer binding, and urea-dependent denaturation.Biochemistry. 1997 May 6;36(18):5311-22. doi: 10.1021/bi961527k. Biochemistry. 1997. PMID: 9154913
-
The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance.J Mol Biol. 1995 Mar 24;247(2):260-80. doi: 10.1006/jmbi.1994.0138. J Mol Biol. 1995. PMID: 7707374
-
Tet repressor induction by tetracycline: a molecular dynamics, continuum electrostatics, and crystallographic study.J Mol Biol. 2008 May 9;378(4):898-912. doi: 10.1016/j.jmb.2008.03.022. Epub 2008 Mar 19. J Mol Biol. 2008. PMID: 18395746
-
The underling mechanism of bacterial TetR/AcrR family transcriptional repressors.Cell Signal. 2013 Jul;25(7):1608-13. doi: 10.1016/j.cellsig.2013.04.003. Epub 2013 Apr 16. Cell Signal. 2013. PMID: 23602932 Review.
Cited by
-
SynPharm and the guide to pharmacology database: A toolset for conferring drug control on engineered proteins.Protein Sci. 2021 Jan;30(1):160-167. doi: 10.1002/pro.3971. Epub 2020 Nov 2. Protein Sci. 2021. PMID: 33047381 Free PMC article.
-
Quorum-sensing regulates biofilm formation in Vibrio scophthalmi.BMC Microbiol. 2012 Dec 3;12:287. doi: 10.1186/1471-2180-12-287. BMC Microbiol. 2012. PMID: 23198796 Free PMC article.
-
Phosphoproteome Dynamics of Streptomyces rimosus during Submerged Growth and Antibiotic Production.mSystems. 2022 Oct 26;7(5):e0019922. doi: 10.1128/msystems.00199-22. Epub 2022 Sep 12. mSystems. 2022. PMID: 36094082 Free PMC article.
-
A quorum sensing-disrupting brominated thiophenone with a promising therapeutic potential to treat luminescent vibriosis.PLoS One. 2012;7(7):e41788. doi: 10.1371/journal.pone.0041788. Epub 2012 Jul 25. PLoS One. 2012. PMID: 22848604 Free PMC article.
-
SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.Proc Natl Acad Sci U S A. 2013 Jun 25;110(26):10586-91. doi: 10.1073/pnas.1221036110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754405 Free PMC article.
References
-
- Ahlert, J., E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet, and J. S. Thorson. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173-1176. - PubMed
-
- Aigle, B., A. Wietzorrek, E. Takano, and M. J. Bibb. 2000. A single amino acid substitution in region 1.2 of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol. Microbiol. 37:995-1004. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

