Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage
- PMID: 15929725
- PMCID: PMC1276948
- DOI: 10.1042/BJ20050379
Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage
Abstract
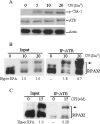
RPA (replication protein A) is an essential factor for DNA DSB (double-strand break) repair and cell cycle checkpoint activation. The 32 kDa subunit of RPA undergoes hyperphosphorylation in response to cellular genotoxic insults. However, the potential involvement of hyperphosphorylated RPA in DSB repair and checkpoint activation remains unclear. Using co-immunoprecipitation assays, we showed that cellular interaction of RPA with two DSB repair factors, Rad51 and Rad52, was predominantly mediated by the hyperphosphorylated species of RPA in cells after UV and camptothecin treatment. Moreover, Rad51 and Rad52 displayed higher affinity for the hyperphosphorylated RPA than native RPA in an in vitro binding assay. Checkpoint kinase ATR (ataxia telangiectasia mutated and Rad3-related) also interacted more efficiently with the hyperphosphorylated RPA than with native RPA following DNA damage. Consistently, immunofluorescence microscopy demonstrated that the hyperphosphorylated RPA was able to co-localize with Rad52 and ATR to form significant nuclear foci in cells. Our results suggest that hyperphosphorylated RPA is preferentially localized to DSB repair and the DNA damage checkpoint complexes in response to DNA damage.
Figures



Similar articles
-
Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates.J Mol Biol. 2009 Jul 3;390(1):45-55. doi: 10.1016/j.jmb.2009.05.009. Epub 2009 May 13. J Mol Biol. 2009. PMID: 19445949
-
RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells.J Mol Biol. 2007 Oct 12;373(1):38-47. doi: 10.1016/j.jmb.2007.07.068. Epub 2007 Aug 15. J Mol Biol. 2007. PMID: 17765923
-
Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments.J Virol. 2009 Jul;83(13):6641-51. doi: 10.1128/JVI.00049-09. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386720 Free PMC article.
-
Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses.J Cell Physiol. 2006 Aug;208(2):267-73. doi: 10.1002/jcp.20622. J Cell Physiol. 2006. PMID: 16523492 Free PMC article. Review.
-
The Intriguing Mystery of RPA Phosphorylation in DNA Double-Strand Break Repair.Genes (Basel). 2024 Jan 27;15(2):167. doi: 10.3390/genes15020167. Genes (Basel). 2024. PMID: 38397158 Free PMC article. Review.
Cited by
-
Cdk5 promotes DNA replication stress checkpoint activation through RPA-32 phosphorylation, and impacts on metastasis free survival in breast cancer patients.Cell Cycle. 2015;14(19):3066-78. doi: 10.1080/15384101.2015.1078020. Cell Cycle. 2015. PMID: 26237679 Free PMC article.
-
DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair.Oncogene. 2013 May 9;32(19):2452-62. doi: 10.1038/onc.2012.257. Epub 2012 Jul 16. Oncogene. 2013. PMID: 22797063 Free PMC article.
-
Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection.J Cell Sci. 2006 Jul 1;119(Pt 13):2695-703. doi: 10.1242/jcs.02981. Epub 2006 Jun 6. J Cell Sci. 2006. PMID: 16757521 Free PMC article.
-
Human replication protein A-Rad52-single-stranded DNA complex: stoichiometry and evidence for strand transfer regulation by phosphorylation.Biochemistry. 2009 Jul 21;48(28):6633-43. doi: 10.1021/bi900564k. Biochemistry. 2009. PMID: 19530647 Free PMC article.
-
Interplay of DNA damage and cell cycle signaling at the level of human replication protein A.DNA Repair (Amst). 2014 Sep;21:12-23. doi: 10.1016/j.dnarep.2014.05.005. Epub 2014 Jun 13. DNA Repair (Amst). 2014. PMID: 25091156 Free PMC article.
References
-
- Wold M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. - PubMed
-
- Iftode C., Daniely Y., Borowiec J. A. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. - PubMed
-
- Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPAssDNA complexes. Science. 2003;300:1542–1548. - PubMed
-
- Binz S. K., Sheehan A. M., Wold M. S. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

