Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair
- PMID: 15923609
- PMCID: PMC1140608
- DOI: 10.1128/MCB.25.12.4903-4913.2005
Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair
Abstract
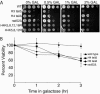
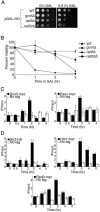
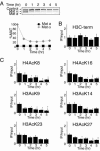
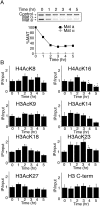
Many recent studies have demonstrated recruitment of chromatin-modifying enzymes to double-strand breaks. Instead, we wanted to examine chromatin modifications during the repair of these double-strand breaks. We show that homologous recombination triggers the acetylation of N-terminal lysines on histones H3 and H4 flanking a double-strand break, followed by deacetylation of H3 and H4. Consistent with a requirement for acetylation and deacetylation during homologous recombination, Saccharomyces cerevisiae with substitutions of the acetylatable lysines of histone H4, deleted for the N-terminal tail of histone H3 or H4, deleted for the histone acetyltransferase GCN5 gene or the histone deacetylase RPD3 gene, shows inviability following induction of an HO lesion that is repaired primarily by homologous recombination. Furthermore, the histone acetyltransferases Gcn5 and Esa1 and the histone deacetylases Rpd3, Sir2, and Hst1 are recruited to the HO lesion during homologous recombinational repair. We have also observed a distinct pattern of histone deacetylation at the donor locus during homologous recombination. Our results demonstrate that dynamic changes in histone acetylation accompany homologous recombination and that the ability to modulate histone acetylation is essential for viability following homologous recombination.
Figures

Similar articles
-
Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4.Mol Cell Biol. 2005 Sep;25(18):8179-90. doi: 10.1128/MCB.25.18.8179-8190.2005. Mol Cell Biol. 2005. PMID: 16135807 Free PMC article.
-
Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair.Nature. 2002 Sep 26;419(6905):411-5. doi: 10.1038/nature01035. Nature. 2002. PMID: 12353039
-
Control of replication initiation by the Sum1/Rfm1/Hst1 histone deacetylase.BMC Mol Biol. 2008 Nov 6;9:100. doi: 10.1186/1471-2199-9-100. BMC Mol Biol. 2008. PMID: 18990212 Free PMC article.
-
Histone acetylation and deacetylation in yeast.Nat Rev Mol Cell Biol. 2003 Apr;4(4):276-84. doi: 10.1038/nrm1075. Nat Rev Mol Cell Biol. 2003. PMID: 12671650 Review.
-
A SAGA of histone acetylation and gene expression.Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available.
Cited by
-
SIRT1 and LSD1 competitively regulate KU70 functions in DNA repair and mutation acquisition in cancer cells.Oncotarget. 2016 Aug 2;7(31):50195-50214. doi: 10.18632/oncotarget.10328. Oncotarget. 2016. PMID: 27384990 Free PMC article.
-
Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus.FEBS Open Bio. 2021 Sep;11(9):2418-2440. doi: 10.1002/2211-5463.13176. Epub 2021 May 19. FEBS Open Bio. 2021. PMID: 33938164 Free PMC article. Review.
-
Gene repression in S. cerevisiae-looking beyond Sir-dependent gene silencing.Curr Genet. 2021 Feb;67(1):3-17. doi: 10.1007/s00294-020-01114-7. Epub 2020 Oct 10. Curr Genet. 2021. PMID: 33037902 Review.
-
Functional Roles of Acetylated Histone Marks at Mouse Meiotic Recombination Hot Spots.Mol Cell Biol. 2017 Jan 19;37(3):e00942-15. doi: 10.1128/MCB.00942-15. Print 2017 Feb 1. Mol Cell Biol. 2017. PMID: 27821479 Free PMC article.
-
Histone modifications: crucial elements for damage response and chromatin restoration.J Cell Physiol. 2010 May;223(2):283-8. doi: 10.1002/jcp.22060. J Cell Physiol. 2010. PMID: 20112283 Free PMC article. Review.
References
-
- Anderson, J. D., P. T. Lowary, and J. Widom. 2001. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 307:977-985. - PubMed
-
- Barlev, N. A., V. Poltoratsky, T. Owen-Hughes, C. Ying, L. Liu, J. L. Workman, and S. L. Berger. 1998. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol. 18:1349-1358. - PMC - PubMed
-
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. - PubMed
-
- Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson, D. E. Hill, and M. Vidal. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295:127-131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
