GATA-1 forms distinct activating and repressive complexes in erythroid cells
- PMID: 15920471
- PMCID: PMC1173143
- DOI: 10.1038/sj.emboj.7600702
GATA-1 forms distinct activating and repressive complexes in erythroid cells
Abstract
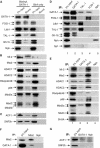
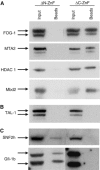
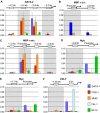
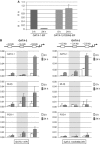
GATA-1 is essential for the generation of the erythroid, megakaryocytic, eosinophilic and mast cell lineages. It acts as an activator and repressor of different target genes, for example, in erythroid cells it represses cell proliferation and early hematopoietic genes while activating erythroid genes, yet it is not clear how both of these functions are mediated. Using a biotinylation tagging/proteomics approach in erythroid cells, we describe distinct GATA-1 interactions with the essential hematopoietic factor Gfi-1b, the repressive MeCP1 complex and the chromatin remodeling ACF/WCRF complex, in addition to the known GATA-1/FOG-1 and GATA-1/TAL-1 complexes. Importantly, we show that FOG-1 mediates GATA-1 interactions with the MeCP1 complex, thus providing an explanation for the overlapping functions of these two factors in erythropoiesis. We also show that subsets of GATA-1 gene targets are bound in vivo by distinct complexes, thus linking specific GATA-1 partners to distinct aspects of its functions. Based on these findings, we suggest a model for the different roles of GATA-1 in erythroid differentiation.
Figures


Similar articles
-
Isolation and characterization of hematopoietic transcription factor complexes by in vivo biotinylation tagging and mass spectrometry.Ann N Y Acad Sci. 2005;1054:55-67. doi: 10.1196/annals.1345.008. Ann N Y Acad Sci. 2005. PMID: 16339652 Review.
-
FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1.EMBO J. 2005 Jul 6;24(13):2367-78. doi: 10.1038/sj.emboj.7600703. Epub 2005 May 26. EMBO J. 2005. PMID: 15920470 Free PMC article.
-
Distinct and overlapping DNMT1 interactions with multiple transcription factors in erythroid cells: Evidence for co-repressor functions.Biochim Biophys Acta. 2016 Dec;1859(12):1515-1526. doi: 10.1016/j.bbagrm.2016.09.007. Epub 2016 Sep 28. Biochim Biophys Acta. 2016. PMID: 27693117
-
GATA-and SP1-binding sites are required for the full activity of the tissue-specific promoter of the tal-1 gene.Oncogene. 1994 Sep;9(9):2623-32. Oncogene. 1994. PMID: 8058326
-
Transcriptional regulation of erythropoiesis: an affair involving multiple partners.Oncogene. 2002 May 13;21(21):3368-76. doi: 10.1038/sj.onc.1205326. Oncogene. 2002. PMID: 12032775 Review.
Cited by
-
Progenitor stage-specific activity of a cis-acting double GATA motif for Gata1 gene expression.Mol Cell Biol. 2015 Mar;35(5):805-15. doi: 10.1128/MCB.01011-14. Epub 2014 Dec 22. Mol Cell Biol. 2015. PMID: 25535330 Free PMC article.
-
FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis.EMBO J. 2010 Jan 20;29(2):457-68. doi: 10.1038/emboj.2009.368. Epub 2009 Dec 10. EMBO J. 2010. PMID: 20010697 Free PMC article.
-
Sumoylation regulates interaction of FOG1 with C-terminal-binding protein (CTBP).J Biol Chem. 2010 Sep 3;285(36):28064-75. doi: 10.1074/jbc.M109.096909. Epub 2010 Jun 28. J Biol Chem. 2010. PMID: 20587419 Free PMC article.
-
Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine.Mol Cell Biol. 2006 Dec;26(23):9060-70. doi: 10.1128/MCB.00124-06. Epub 2006 Aug 28. Mol Cell Biol. 2006. PMID: 16940177 Free PMC article.
-
The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation.Genes Dev. 2010 Feb 1;24(3):277-89. doi: 10.1101/gad.551810. Genes Dev. 2010. PMID: 20123907 Free PMC article.
References
-
- Ahringer J (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 - PubMed
-
- Anderson KP, Crable SC, Lingrel JB (1998) Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J Biol Chem 273: 14347–14354 - PubMed
-
- Blobel GA, Weiss MJ (2001) Nuclear factors that regulate erythropoiesis. In Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, Steinberg MH, Forget BG, Higgs DR, Nagel RL (eds) pp 72–94. Cambridge: Cambridge University Press
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

