Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane
- PMID: 15890898
- PMCID: PMC1112135
- DOI: 10.1128/JVI.79.11.6598-6609.2005
Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane
Abstract
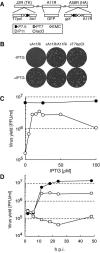
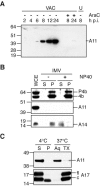
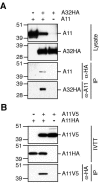
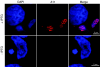
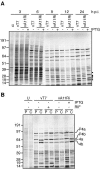
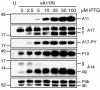
The vaccinia virus A11R gene has orthologs in all known poxvirus genomes, and the A11 protein has been previously reported to interact with the putative DNA packaging protein A32 in a yeast two-hybrid screen. Using antisera raised against A11 peptides, we show that the A11 protein was (i) expressed at late times with an apparent mass of 40 kDa, (ii) not incorporated into virus particles, (iii) phosphorylated independently of the viral F10 kinase, (iv) coimmunoprecipitated with A32, and (v) localized to the viral factory. To determine the role of the A11 protein and test whether it is indeed involved in DNA packaging, we constructed a recombinant vaccinia virus with an inducible A11R gene. This recombinant was dependent on inducer for single-cycle growth and plaque formation. In the absence of inducer, viral late proteins were produced at normal levels, but proteolytic processing and other posttranslational modifications of some proteins were inhibited, suggesting a block in virus particle assembly. Consistent with this observation, electron microscopy of cells infected in the absence of inducer showed virus factories with abnormal electron-dense viroplasms and intermediate density regions associated with membranes and containing the D13 protein. However, no viral membrane crescents, immature virions, or mature virions were produced. The requirement for nonvirion protein A11 in order to make normal viral membranes was an unexpected and exciting finding, since neither the origin of these membranes nor their mechanism of formation in the cytoplasm of infected cells is understood.
Figures



Similar articles
-
Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature.J Virol. 2004 Oct;78(19):10238-48. doi: 10.1128/JVI.78.19.10238-10248.2004. J Virol. 2004. PMID: 15367589 Free PMC article.
-
A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions.Virology. 2004 Dec 20;330(2):447-59. doi: 10.1016/j.virol.2004.10.008. Virology. 2004. PMID: 15567438
-
Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions.J Virol. 2001 Jul;75(13):5752-61. doi: 10.1128/JVI.75.13.5752-5761.2001. J Virol. 2001. PMID: 11390577 Free PMC article.
-
Vaccinia virus motility.Annu Rev Microbiol. 2003;57:323-42. doi: 10.1146/annurev.micro.57.030502.091037. Annu Rev Microbiol. 2003. PMID: 14527282 Review.
-
Vaccinia virus morphogenesis and dissemination.Trends Microbiol. 2008 Oct;16(10):472-9. doi: 10.1016/j.tim.2008.07.009. Epub 2008 Sep 12. Trends Microbiol. 2008. PMID: 18789694 Review.
Cited by
-
Poxvirus membrane biogenesis.Virology. 2015 May;479-480:619-26. doi: 10.1016/j.virol.2015.02.003. Epub 2015 Feb 26. Virology. 2015. PMID: 25728299 Free PMC article. Review.
-
The E6 protein from vaccinia virus is required for the formation of immature virions.Virology. 2010 Apr 10;399(2):201-11. doi: 10.1016/j.virol.2010.01.012. Epub 2010 Feb 8. Virology. 2010. PMID: 20116821 Free PMC article.
-
Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly.J Virol. 2012 May;86(10):5603-13. doi: 10.1128/JVI.00330-12. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398288 Free PMC article.
-
Poxviruses Encode a Reticulon-Like Protein that Promotes Membrane Curvature.Cell Rep. 2016 Mar 8;14(9):2084-2091. doi: 10.1016/j.celrep.2016.01.075. Epub 2016 Feb 25. Cell Rep. 2016. PMID: 26923595 Free PMC article.
-
Association of the vaccinia virus A11 protein with the endoplasmic reticulum and crescent precursors of immature virions.J Virol. 2013 Sep;87(18):10195-206. doi: 10.1128/JVI.01601-13. Epub 2013 Jul 17. J Virol. 2013. PMID: 23864611 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

