Genetic and antigenic variability of human respiratory syncytial virus (groups a and b) isolated over seven consecutive seasons in Argentina (1995 to 2001)
- PMID: 15872254
- PMCID: PMC1153737
- DOI: 10.1128/JCM.43.5.2266-2273.2005
Genetic and antigenic variability of human respiratory syncytial virus (groups a and b) isolated over seven consecutive seasons in Argentina (1995 to 2001)
Abstract
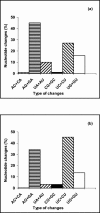
The genetic and antigenic variability of human respiratory syncytial virus (HRSV) strains isolated in Buenos Aires from 1995 to 2001 was evaluated by partial nucleotide sequencing of the G gene and enzyme-linked immunosorbent assay analysis with anti-G monoclonal antibodies. Phylogenetic analyses showed that 37 group A strains clustered into five genotypes, whereas 20 group B strains clustered into three genotypes. Group A showed more genetic variability than group B. A close correlation between genotypes and antigenic patterns was observed. Changes detected in the G protein of viruses from both groups included (i) amino acid substitutions and(ii) differences in protein length due to either changes in stop codon usage or sequence duplications. Three B strains from 1999 exhibited a duplication of 20 amino acids, while one B strain from 2001 had 2 amino acids duplicated. The comparison among Argentinean HRSV strains and viruses isolated in other geographical areas during different epidemics is discussed.
Figures



Similar articles
-
Antigenic and genetic variability of human respiratory syncytial viruses (group A) isolated in Uruguay and Argentina: 1993-2001.J Med Virol. 2003 Oct;71(2):305-12. doi: 10.1002/jmv.10484. J Med Virol. 2003. PMID: 12938207
-
Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons.J Clin Microbiol. 2007 Sep;45(9):3022-30. doi: 10.1128/JCM.00339-07. Epub 2007 Jul 3. J Clin Microbiol. 2007. PMID: 17609323 Free PMC article.
-
Intragroup antigenic diversity of human respiratory syncytial virus (group A) isolated in Argentina and Chile.J Med Virol. 2005 Oct;77(2):311-6. doi: 10.1002/jmv.20456. J Med Virol. 2005. PMID: 16121383
-
Antigenic and genetic variation in human respiratory syncytial virus.Pediatr Infect Dis J. 2004 Jan;23(1 Suppl):S19-24. doi: 10.1097/01.inf.0000108189.87181.7c. Pediatr Infect Dis J. 2004. PMID: 14730266 Review.
-
Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985-2001--a review.Mem Inst Oswaldo Cruz. 2005 May;100(3):221-30. doi: 10.1590/s0074-02762005000300001. Epub 2005 Aug 15. Mem Inst Oswaldo Cruz. 2005. PMID: 16113858 Review.
Cited by
-
Genetic variability of respiratory syncytial viruses (RSV) prevalent in Southwestern China from 2006 to 2009: emergence of subgroup B and A RSV as dominant strains.J Clin Microbiol. 2010 Apr;48(4):1201-7. doi: 10.1128/JCM.02258-09. Epub 2010 Feb 10. J Clin Microbiol. 2010. PMID: 20147636 Free PMC article.
-
Does the viral subtype influence the biennial cycle of respiratory syncytial virus?Virol J. 2009 Sep 7;6:133. doi: 10.1186/1743-422X-6-133. Virol J. 2009. PMID: 19735540 Free PMC article.
-
Novel clinical features of recurrent human respiratory syncytial virus infections.J Med Virol. 2014 Sep;86(9):1629-38. doi: 10.1002/jmv.23809. Epub 2013 Oct 25. J Med Virol. 2014. PMID: 24166209 Free PMC article.
-
Evaluation of respiratory syncytial virus group A and B genotypes among nosocomial and community-acquired pediatric infections in Southern Brazil.Virol J. 2014 Feb 24;11:36. doi: 10.1186/1743-422X-11-36. Virol J. 2014. PMID: 24564922 Free PMC article.
-
Genetic Diversity in the G Protein Gene of Human Respiratory Syncytial Virus among Iranian Children with Acute Respiratory Symptoms.Iran J Pediatr. 2011 Mar;21(1):58-64. Iran J Pediatr. 2011. PMID: 23056765 Free PMC article.
References
-
- Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. - PubMed
-
- Baumeister, E. G., D. S. Hunicken, and V. L. Savy. 2003. RSV molecular characterization and specific antibody response in young children with acute lower respiratory infection. J. Clin. Virol. 27:44-51. - PubMed
-
- Blanc, A., A. Delfraro, S. Frabasile, and J. Arbiza. 2005. Genotypes of respiratory syncytial virus group B identified in Uruguay. Arch. Virol. 150:603-609. - PubMed
-
- Cane, P. A., D. A. Matthews, and C. R. Pringle. 1991. Identification of variable domains of the attachment (G) protein of subgroup A respiratory syncytial viruses. J. Gen. Virol. 72:2091-2096. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

