Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus
- PMID: 15848177
- PMCID: PMC7094623
- DOI: 10.1016/j.febslet.2005.03.039
Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus
Abstract
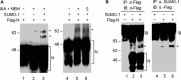
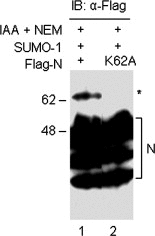
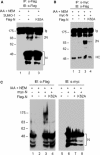
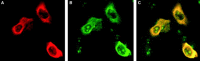
Severe acute respiratory syndrome coronavirus (SARS-CoV) encodes a highly basic nucleocapsid (N) protein of 422 amino acids. Similar to other coronavirus N proteins, SARS-CoV N protein is predicted to be phosphorylated and may contain nuclear localization signals, serine/arginine-rich motif, RNA binding domain and regions responsible for self-association and homo-oligomerization. In this study, we demonstrate that the protein is posttranslationally modified by covalent attachment to the small ubiquitin-like modifier. The major sumoylation site was mapped to the (62)lysine residue of the N protein. Further expression and characterization of wild type N protein and K62A mutant reveal that sumoylation of the N protein drastically promotes its homo-oligomerization, and plays certain roles in the N protein-mediated interference of host cell division. This is the first report showing that a coronavirus N protein undergoes posttranslational modification by sumoylation, and the functional implication of this modification in the formation of coronavirus ribouncleoprotein complex, virion assembly and virus-host interactions.
Figures


Similar articles
-
SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system.J Med Virol. 2006 Nov;78(11):1365-73. doi: 10.1002/jmv.20707. J Med Virol. 2006. PMID: 16998888 Free PMC article.
-
Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus by interaction with Ubc9.Adv Exp Med Biol. 2006;581:121-6. doi: 10.1007/978-0-387-33012-9_21. Adv Exp Med Biol. 2006. PMID: 17037517 Free PMC article. No abstract available.
-
The nucleocapsid protein of SARS-associated coronavirus inhibits B23 phosphorylation.Biochem Biophys Res Commun. 2008 May 2;369(2):287-91. doi: 10.1016/j.bbrc.2008.01.096. Epub 2008 Feb 1. Biochem Biophys Res Commun. 2008. PMID: 18243139 Free PMC article.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
-
The SARS-CoV nucleocapsid protein: a protein with multifarious activities.Infect Genet Evol. 2008 Jul;8(4):397-405. doi: 10.1016/j.meegid.2007.07.004. Epub 2007 Jul 20. Infect Genet Evol. 2008. PMID: 17881296 Free PMC article. Review.
Cited by
-
A viral assembly inhibitor blocks SARS-CoV-2 replication in airway epithelial cells.Commun Biol. 2024 Apr 22;7(1):486. doi: 10.1038/s42003-024-06130-8. Commun Biol. 2024. PMID: 38649430 Free PMC article.
-
TRIM28-mediated nucleocapsid protein SUMOylation enhances SARS-CoV-2 virulence.Nat Commun. 2024 Jan 4;15(1):244. doi: 10.1038/s41467-023-44502-6. Nat Commun. 2024. PMID: 38172120 Free PMC article.
-
The emerging roles of SUMOylation in pulmonary diseases.Mol Med. 2023 Sep 5;29(1):119. doi: 10.1186/s10020-023-00719-1. Mol Med. 2023. PMID: 37670258 Free PMC article. Review.
-
Human Post-Translational SUMOylation Modification of SARS-CoV-2 Nucleocapsid Protein Enhances Its Interaction Affinity with Itself and Plays a Critical Role in Its Nuclear Translocation.Viruses. 2023 Jul 21;15(7):1600. doi: 10.3390/v15071600. Viruses. 2023. PMID: 37515286 Free PMC article.
-
A Novel Viral Assembly Inhibitor Blocks SARS-CoV-2 Replication in Airway Epithelial Cells.Res Sq [Preprint]. 2023 May 17:rs.3.rs-2887435. doi: 10.21203/rs.3.rs-2887435/v1. Res Sq. 2023. Update in: Commun Biol. 2024 Apr 22;7(1):486. doi: 10.1038/s42003-024-06130-8. PMID: 37292622 Free PMC article. Updated. Preprint.
References
-
- Chang L.K., Lee Y.H., Cheng T.S., Hong Y.R., Lu P.J., Wang J.J., Wang W.H., Kuo C.W., Li S.S., Liu S.T., Posttranslational modification of Rta of Epstein–Barr virus by SUMO-1. J. Biol. Chem., 279, (2004), 38803– 38812. - PubMed
-
- Davies H.A., Dourmashkin R.R., MacNaughton R., Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol., 53, (1981), 67– 74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

