Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa
- PMID: 15831483
- PMCID: PMC1084287
- DOI: 10.1128/MCB.25.9.3793-3801.2005
Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa
Abstract
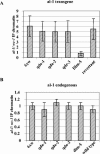
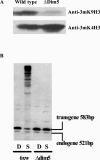
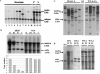
In Neurospora crassa, the introduction of a transgene can lead to small interfering RNA (siRNA)-mediated posttranscriptional gene silencing (PTGS) of homologous genes. siRNAs can also guide locus-specific methylation of Lys9 of histone H3 (Lys9H3) in Schizosaccharomyces pombe. Here we tested the hypothesis that transgenically derived siRNAs may contemporaneously both activate the PTGS mechanism and induce chromatin modifications at the transgene and the homologous endogenous gene. We carried out chromatin immunoprecipitation using a previously characterized albino-1 (al-1) silenced strain but detected no alterations in the pattern of histone modifications at the endogenous al-1 locus, suggesting that siRNAs produced from the transgenic locus do not trigger modifications in trans of those histones tested. Instead, we found that the transgenic locus was hypermethylated at Lys9H3 in our silenced strain and remained hypermethylated in the quelling defective mutants (qde), further demonstrating that the PTGS machinery is dispensable for Lys9H3 methylation. However, we found that a mutant in the histone Lys9H3 methyltransferase dim-5 was unable to maintain PTGS, with transgenic copies being rapidly lost, resulting in reversion of the silenced phenotype. These results indicate that the defect in PTGS of the Deltadim-5 strain is due to the inability to maintain the transgene in tandem, suggesting a role for DIM-5 in stabilizing such repeated sequences. We conclude that in Neurospora, siRNAs produced from the transgenic locus are used in the RNA-induced silencing complex-mediated PTGS pathway and do not communicate with an RNAi-induced initiation of transcriptional gene silencing complex to effect chromatin-based silencing.
Figures

Similar articles
-
RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa.Nucleic Acids Res. 2004 Aug 9;32(14):4237-43. doi: 10.1093/nar/gkh764. Print 2004. Nucleic Acids Res. 2004. PMID: 15302921 Free PMC article.
-
A histone H3 methyltransferase controls DNA methylation in Neurospora crassa.Nature. 2001 Nov 15;414(6861):277-83. doi: 10.1038/35104508. Nature. 2001. PMID: 11713521
-
Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa.Nat Genet. 2003 May;34(1):75-9. doi: 10.1038/ng1143. Nat Genet. 2003. PMID: 12679815
-
Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa.Curr Opin Microbiol. 2007 Apr;10(2):199-203. doi: 10.1016/j.mib.2007.03.016. Epub 2007 Mar 28. Curr Opin Microbiol. 2007. PMID: 17395524 Review.
-
The many faces of histone lysine methylation.Curr Opin Cell Biol. 2002 Jun;14(3):286-98. doi: 10.1016/s0955-0674(02)00335-6. Curr Opin Cell Biol. 2002. PMID: 12067650 Review.
Cited by
-
The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells.Mol Cell Biol. 2006 Jun;26(11):4028-40. doi: 10.1128/MCB.02189-05. Mol Cell Biol. 2006. PMID: 16705157 Free PMC article.
-
Neurospora crassa, a model system for epigenetics research.Cold Spring Harb Perspect Biol. 2013 Oct 1;5(10):a017921. doi: 10.1101/cshperspect.a017921. Cold Spring Harb Perspect Biol. 2013. PMID: 24086046 Free PMC article. Review.
-
A double-stranded-RNA response program important for RNA interference efficiency.Mol Cell Biol. 2007 Jun;27(11):3995-4005. doi: 10.1128/MCB.00186-07. Epub 2007 Mar 19. Mol Cell Biol. 2007. PMID: 17371837 Free PMC article.
-
Copy number-dependent DNA methylation of the Pyricularia oryzae MAGGY retrotransposon is triggered by DNA damage.Commun Biol. 2021 Mar 19;4(1):351. doi: 10.1038/s42003-021-01836-5. Commun Biol. 2021. PMID: 33742058 Free PMC article.
-
Evolution and diversification of RNA silencing proteins in fungi.J Mol Evol. 2006 Jul;63(1):127-35. doi: 10.1007/s00239-005-0257-2. Epub 2006 Jun 16. J Mol Evol. 2006. PMID: 16786437
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
