SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4
- PMID: 15831457
- PMCID: PMC1084305
- DOI: 10.1128/MCB.25.9.3506-3518.2005
SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4
Abstract
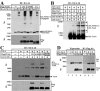
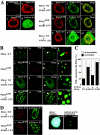
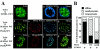
We have previously shown that modification of Tcf-4, a transcription factor in the Wnt pathway, with SUMO by PIASy, a SUMO E3 ligase, enhances its transcriptional activity. Since PIASy itself was also modified with SUMO-1, we studied the role of sumoylation of PIASy in the regulation of Tcf-4. Lys(35) was found to be a sumoylation site of PIASy. PIASy(K35R), in which Lys(35) was mutated to Arg, did not enhance sumoylation of Tcf-4, although this PIASy mutant did not lose the ligase activity of sumoylation for other proteins. Wild-type PIASy and PIASy(K35R) showed a distinct distribution in the nucleus, although both were colocalized with Tcf-4. Promyelocytic leukemia protein, which is involved in transcriptional regulation, was associated with PIASy(K35R) more frequently than wild-type PIASy in the nucleus. PIASy(K35R) could not stimulate the transcriptional activity of Tcf-4 under the conditions in which wild-type PIASy enhanced it. Conjugation of SUMO-1 to the amino terminus of PIASy(K35R) neither enhanced sumoylation of Tcf-4 nor stimulated the transcriptional activity of Tcf-4. These results suggest that sumoylation of Lys(35) in PIASy determines the nuclear localization of PIASy and that it is necessary for PIASy-dependent sumoylation and transcriptional activation of Tcf-4.
Figures




Similar articles
-
The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification.Mol Cell Biol. 2005 Mar;25(5):1879-90. doi: 10.1128/MCB.25.5.1879-1890.2005. Mol Cell Biol. 2005. PMID: 15713642 Free PMC article.
-
Identification of a new small ubiquitin-like modifier (SUMO)-interacting motif in the E3 ligase PIASy.J Biol Chem. 2017 Jun 16;292(24):10230-10238. doi: 10.1074/jbc.M117.789982. Epub 2017 Apr 28. J Biol Chem. 2017. PMID: 28455449 Free PMC article.
-
Transactivation properties of c-Myb are critically dependent on two SUMO-1 acceptor sites that are conjugated in a PIASy enhanced manner.Eur J Biochem. 2003 Mar;270(6):1338-48. doi: 10.1046/j.1432-1033.2003.03504.x. Eur J Biochem. 2003. PMID: 12631292
-
Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival.Mol Carcinog. 2007 Aug;46(8):585-90. doi: 10.1002/mc.20344. Mol Carcinog. 2007. PMID: 17626251 Review.
-
PIAS/SUMO: new partners in transcriptional regulation.Cell Mol Life Sci. 2003 Dec;60(12):2561-74. doi: 10.1007/s00018-003-3129-1. Cell Mol Life Sci. 2003. PMID: 14685683 Free PMC article. Review.
Cited by
-
High throughput genomic screen identifies multiple factors that promote cooperative Wnt signaling.PLoS One. 2013;8(1):e55782. doi: 10.1371/journal.pone.0055782. Epub 2013 Jan 31. PLoS One. 2013. PMID: 23383281 Free PMC article.
-
Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes.Plant Physiol. 2009 Dec;151(4):1930-42. doi: 10.1104/pp.109.143719. Epub 2009 Oct 16. Plant Physiol. 2009. PMID: 19837819 Free PMC article.
-
Functional mimicry of the acetylated C-terminal tail of p53 by a SUMO-1 acetylated domain, SAD.J Cell Physiol. 2010 Nov;225(2):371-84. doi: 10.1002/jcp.22224. J Cell Physiol. 2010. PMID: 20458745 Free PMC article.
-
Aberrant sumoylation signaling evoked by reactive oxygen species impairs protective function of Prdx6 by destabilization and repression of its transcription.FEBS J. 2014 Aug;281(15):3357-81. doi: 10.1111/febs.12866. Epub 2014 Jul 1. FEBS J. 2014. PMID: 24910119 Free PMC article.
-
Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2.Nucleus. 2012 Jul 1;3(4):349-58. doi: 10.4161/nucl.20822. Epub 2012 Jun 12. Nucleus. 2012. PMID: 22688647 Free PMC article.
References
-
- Abmayr, S. M., and J. L. Workman. 1989. Preparation of nuclear and cytoplasmic extracts from mammalian cells, p. 12.1.1-12.1.9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley, New York, N.Y. - PubMed
-
- Bienz, M., and H. Clevers. 2000. Linking colorectal cancer to Wnt signaling. Cell 103:311-320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
