Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3' boundary sequence spanning 46 kilobases
- PMID: 15798207
- PMCID: PMC1069589
- DOI: 10.1128/MCB.25.8.3220-3231.2005
Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3' boundary sequence spanning 46 kilobases
Abstract
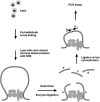
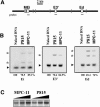
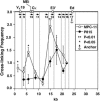
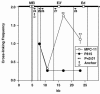
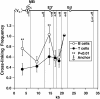
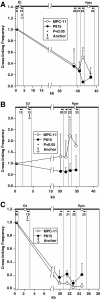
The mouse immunoglobulin kappa (Igkappa) gene contains an intronic enhancer and two enhancers downstream of its transcription unit. Using chromosome conformation capture technology, we demonstrate that rearranged and actively transcribed Igkappa alleles in MPC-11 plasmacytoma cells exhibit mutual interactions over 22 kb between these three enhancers and Vkappa gene promoters. In addition, the 5' region of the active transcription unit exhibits a continuum of interactions with downstream chromatin segments. We also observe interactions between Ei and E3' with 3' boundary sequences 24 kb downstream of Ed, adjacent to a neighboring housekeeping gene. Very similar interactions between the enhancers are also exhibited by normal B cells isolated from mouse splenic tissue but not by germ line transcriptionally inactive alleles of T cells or P815 mastocytoma cells, which exhibit a seemingly linear chromatin organization. These results fit a looping mechanism for enhancer function like in the beta-globin locus and suggest a dynamic modulation of the spatial organization of the active Igkappa locus. Chromatin immunoprecipitation experiments reveal that the interacting Igkappa gene cis-acting sequences are associated with AP-4, E47, and p65NF-kappaB, potential protein candidates that may be responsible for initiating and/or maintaining the formation of these higher-order complexes. However, S107 plasmacytoma cells that lack NF-kappaB still exhibit mutual interactions between the Igkappa gene enhancers.
Figures
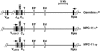


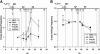

Similar articles
-
The Igκ gene enhancers, E3' and Ed, are essential for triggering transcription.J Immunol. 2010 Dec 15;185(12):7544-52. doi: 10.4049/jimmunol.1002665. Epub 2010 Nov 12. J Immunol. 2010. PMID: 21076060 Free PMC article.
-
Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin κ Light-Chain Locus.Adv Immunol. 2015;128:183-271. doi: 10.1016/bs.ai.2015.07.004. Epub 2015 Aug 14. Adv Immunol. 2015. PMID: 26477368 Review.
-
YY1 binds to the E3' enhancer and inhibits the expression of the immunoglobulin κ gene via epigenetic modifications.Immunology. 2018 Dec;155(4):491-498. doi: 10.1111/imm.12990. Epub 2018 Aug 29. Immunology. 2018. PMID: 30098214 Free PMC article.
-
E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells.Mol Cell Biol. 2006 Feb;26(3):810-21. doi: 10.1128/MCB.26.3.810-821.2006. Mol Cell Biol. 2006. PMID: 16428437 Free PMC article.
-
Nongenic transcription, gene regulation and action at a distance.J Cell Sci. 2003 Nov 15;116(Pt 22):4483-91. doi: 10.1242/jcs.00819. J Cell Sci. 2003. PMID: 14576342 Review.
Cited by
-
Genome organizing function of SATB1 in tumor progression.Semin Cancer Biol. 2013 Apr;23(2):72-9. doi: 10.1016/j.semcancer.2012.06.009. Epub 2012 Jul 4. Semin Cancer Biol. 2013. PMID: 22771615 Free PMC article. Review.
-
Developmental regulation of chromatin conformation by Hox proteins in Drosophila.Cell Rep. 2012 Apr 19;1(4):350-9. doi: 10.1016/j.celrep.2012.03.003. Cell Rep. 2012. PMID: 22523743 Free PMC article.
-
Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements.J Phys Chem B. 2016 Aug 25;120(33):8642-53. doi: 10.1021/acs.jpcb.6b03197. Epub 2016 Jun 16. J Phys Chem B. 2016. PMID: 27218881 Free PMC article.
-
Spatial configuration of the chicken alpha-globin gene domain: immature and active chromatin hubs.Nucleic Acids Res. 2008 Aug;36(14):4629-40. doi: 10.1093/nar/gkn429. Epub 2008 Jul 11. Nucleic Acids Res. 2008. PMID: 18621783 Free PMC article.
-
Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes.Genes Dev. 2008 Aug 15;22(16):2204-14. doi: 10.1101/gad.1675908. Genes Dev. 2008. PMID: 18708579 Free PMC article.
References
-
- Atchison, M. L., and R. P. Perry. 1987. The role of the κ enhancer and its binding factor NF-kB in the developmental regulation of κ gene transcription. Cell 48:121-128. - PubMed
-
- Bain, G., E. C. R. Maandag, H. P. J. te Riele, A. J. Freeney, A. Sheehy, M. Schlissel, S. A. Shinton, R. R. Hardy, and C. Murre. 1997. Both E12 and E47 allow commitment to the B cell lineage. Immunity 6:145-154. - PubMed
-
- Baxter, J., M. Merkenschlager, and A. G. Fisher. 2002. Nuclear organization and gene expression. Curr. Opin. Cell Biol. 14:372-376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
