Impairment of replication fork progression mediates RNA polII transcription-associated recombination
- PMID: 15775982
- PMCID: PMC556405
- DOI: 10.1038/sj.emboj.7600602
Impairment of replication fork progression mediates RNA polII transcription-associated recombination
Abstract
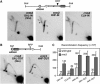
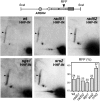
Homologous recombination safeguards genome integrity, but it can also cause genome instability of important consequences for cell proliferation and organism development. Transcription induces recombination, as shown in prokaryotes and eukaryotes for both spontaneous and developmentally regulated events such as those responsible for immunoglobulin class switching. Deciphering the molecular basis of transcription-associated recombination (TAR) is important in understanding genome instability. Using novel plasmid-borne recombination constructs in Saccharomyces cerevisiae, we show that RNA polymerase II (RNAPII) transcription induces recombination by impairing replication fork progression. RNAPII transcription concomitant to head-on oncoming replication causes a replication fork pause (RFP) that is linked to a significant increase in recombination. However, transcription that is codirectional with replication has little effect on replication fork progression and recombination. Transcription occurring in the absence of replication does not affect either recombination or replication fork progression. The Rrm3 helicase, which is required for replication fork progression through nucleoprotein complexes, facilitates replication through the transcription-dependent RFP site and reduces recombination. Therefore, our work provides evidence that one mechanism responsible for TAR is RNAP-mediated replication impairment.
Figures
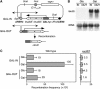
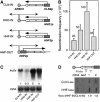
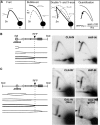
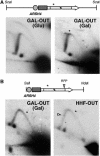

Similar articles
-
Stimulation of direct-repeat recombination by RNA polymerase III transcription.DNA Repair (Amst). 2009 May 1;8(5):620-6. doi: 10.1016/j.dnarep.2008.12.010. Epub 2009 Jan 24. DNA Repair (Amst). 2009. PMID: 19168400
-
Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex.Mol Cell Biol. 2006 Apr;26(8):3327-34. doi: 10.1128/MCB.26.8.3327-3334.2006. Mol Cell Biol. 2006. PMID: 16581804 Free PMC article.
-
RNA polymerase II contributes to preventing transcription-mediated replication fork stalls.EMBO J. 2015 Jan 13;34(2):236-50. doi: 10.15252/embj.201488544. Epub 2014 Dec 1. EMBO J. 2015. PMID: 25452497 Free PMC article.
-
Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination.Mutagenesis. 2009 May;24(3):203-10. doi: 10.1093/mutage/gen072. Epub 2009 Jan 12. Mutagenesis. 2009. PMID: 19139058 Review.
-
Stalled replication forks: making ends meet for recognition and stabilization.Bioessays. 2010 Aug;32(8):687-97. doi: 10.1002/bies.200900196. Bioessays. 2010. PMID: 20658707 Review.
Cited by
-
Breaking the paradigm: early insights from mammalian DNA breakomes.FEBS J. 2022 May;289(9):2409-2428. doi: 10.1111/febs.15849. Epub 2021 May 1. FEBS J. 2022. PMID: 33792193 Free PMC article. Review.
-
Unscheduled DNA synthesis leads to elevated uracil residues at highly transcribed genomic loci in Saccharomyces cerevisiae.PLoS Genet. 2018 Jul 17;14(7):e1007516. doi: 10.1371/journal.pgen.1007516. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30016327 Free PMC article.
-
Ribosome Collisions Result in +1 Frameshifting in the Absence of No-Go Decay.Cell Rep. 2019 Aug 13;28(7):1679-1689.e4. doi: 10.1016/j.celrep.2019.07.046. Cell Rep. 2019. PMID: 31412239 Free PMC article.
-
Transcription-induced formation of extrachromosomal DNA during yeast ageing.PLoS Biol. 2019 Dec 3;17(12):e3000471. doi: 10.1371/journal.pbio.3000471. eCollection 2019 Dec. PLoS Biol. 2019. PMID: 31794573 Free PMC article.
-
Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication.DNA Repair (Amst). 2007 Sep 1;6(9):1285-96. doi: 10.1016/j.dnarep.2007.02.023. Epub 2007 Mar 29. DNA Repair (Amst). 2007. PMID: 17398168 Free PMC article.
References
-
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 - PubMed
-
- Brewer BJ, Fangman WL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 - PubMed
-
- Casper AM, Nghiem P, Arlt MF, Glover TW (2002) ATR regulates fragile site stability. Cell 111: 779–789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

