Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression
- PMID: 15774026
- PMCID: PMC1088944
- DOI: 10.1186/gb-2005-6-3-r25
Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression
Abstract
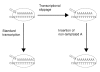
Background: Transcription slippage occurs on certain patterns of repeat mononucleotides, resulting in synthesis of a heterogeneous population of mRNAs. Individual mRNA molecules within this population differ in the number of nucleotides they contain that are not specified by the template. When transcriptional slippage occurs in a coding sequence, translation of the resulting mRNAs yields more than one protein product. Except where the products of the resulting mRNAs have distinct functions, transcription slippage occurring in a coding region is expected to be disadvantageous. This probably leads to selection against most slippage-prone sequences in coding regions.
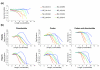
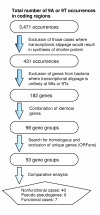
Results: To find a length at which such selection is evident, we analyzed the distribution of repetitive runs of A and T of different lengths in 108 bacterial genomes. This length varies significantly among different bacteria, but in a large proportion of available genomes corresponds to nine nucleotides. Comparative sequence analysis of these genomes was used to identify occurrences of 9A and 9T transcriptional slippage-prone sequences used for gene expression.
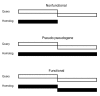
Conclusions: IS element genes are the largest group found to exploit this phenomenon. A number of genes with disrupted open reading frames (ORFs) have slippage-prone sequences at which transcriptional slippage would result in uninterrupted ORF restoration at the mRNA level. The ability of such genes to encode functional full-length protein products brings into question their annotation as pseudogenes and in these cases is pertinent to the significance of the term 'authentic frameshift' frequently assigned to such genes.
Figures
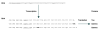

Similar articles
-
Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts.Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14934-9. doi: 10.1073/pnas.0806554105. Epub 2008 Sep 24. Proc Natl Acad Sci U S A. 2008. PMID: 18815381 Free PMC article.
-
Characterization of 954 bovine full-CDS cDNA sequences.BMC Genomics. 2005 Nov 23;6:166. doi: 10.1186/1471-2164-6-166. BMC Genomics. 2005. PMID: 16305752 Free PMC article.
-
Sequences of 3' end of genome and of 5' end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5' leader and bodies of seven subgenomic mRNAs.J Gen Virol. 1993 Apr;74 ( Pt 4):643-59. doi: 10.1099/0022-1317-74-4-643. J Gen Virol. 1993. PMID: 8385693
-
The nature and dynamics of bacterial genomes.Science. 2006 Mar 24;311(5768):1730-3. doi: 10.1126/science.1119966. Science. 2006. PMID: 16556833 Review.
-
When one is better than two: RNA with dual functions.Biochimie. 2011 Apr;93(4):633-44. doi: 10.1016/j.biochi.2010.11.004. Epub 2010 Nov 24. Biochimie. 2011. PMID: 21111023 Review.
Cited by
-
Avoidance of long mononucleotide repeats in codon pair usage.Genetics. 2010 Nov;186(3):1077-84. doi: 10.1534/genetics.110.121137. Epub 2010 Aug 30. Genetics. 2010. PMID: 20805553 Free PMC article.
-
Specific reverse transcriptase slippage at the HIV ribosomal frameshift sequence: potential implications for modulation of GagPol synthesis.Nucleic Acids Res. 2017 Sep 29;45(17):10156-10167. doi: 10.1093/nar/gkx690. Nucleic Acids Res. 2017. PMID: 28973470 Free PMC article.
-
Correction of frameshift mutations in the atpB gene by translational recoding in chloroplasts of Oenothera and tobacco.Plant Cell. 2021 Jul 2;33(5):1682-1705. doi: 10.1093/plcell/koab050. Plant Cell. 2021. PMID: 33561268 Free PMC article.
-
Productive mRNA stem loop-mediated transcriptional slippage: Crucial features in common with intrinsic terminators.Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):E1984-93. doi: 10.1073/pnas.1418384112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848054 Free PMC article.
-
CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut.J Bacteriol. 2007 Mar;189(5):1856-65. doi: 10.1128/JB.01427-06. Epub 2006 Dec 15. J Bacteriol. 2007. PMID: 17172331 Free PMC article.
References
-
- Liu C, Heath LS, Turnbough CL., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–2912. - PubMed
-
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

