The first steps of transposable elements invasion: parasitic strategy vs. genetic drift
- PMID: 15731520
- PMCID: PMC1449084
- DOI: 10.1534/genetics.104.031211
The first steps of transposable elements invasion: parasitic strategy vs. genetic drift
Abstract
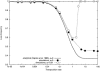
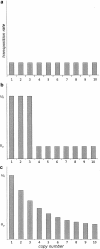
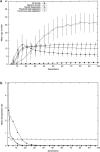
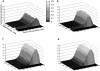
Transposable elements are often considered as selfish DNA sequences able to invade the genome of their host species. Their evolutive dynamics are complex, due to the interaction between their intrinsic amplification capacity, selection at the host level, transposition regulation, and genetic drift. Here, we propose modeling the first steps of TE invasion, i.e., just after a horizontal transfer, when a single copy is present in the genome of one individual. If the element has a constant transposition rate, it will disappear in most cases: the elements with low-transposition rate are frequently lost through genetic drift, while those with high-transposition rate may amplify, leading to the sterility of their host. Elements whose transposition rate is regulated are able to successfully invade the populations, thanks to an initial transposition burst followed by a strong limitation of their activity. Self-regulation or hybrid dysgenesis may thus represent some genome-invasion parasitic strategies.
Figures

Similar articles
-
Horizontal Transfer Can Drive a Greater Transposable Element Load in Large Populations.J Hered. 2017 Jan;108(1):36-44. doi: 10.1093/jhered/esw050. Epub 2016 Aug 24. J Hered. 2017. PMID: 27558983
-
The dynamics of transposable elements in structured populations.Genetics. 2005 Jan;169(1):467-74. doi: 10.1534/genetics.104.032243. Epub 2004 Sep 30. Genetics. 2005. PMID: 15466430 Free PMC article.
-
Long-term evolution of transposable elements.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19375-80. doi: 10.1073/pnas.0705238104. Epub 2007 Nov 26. Proc Natl Acad Sci U S A. 2007. PMID: 18040048 Free PMC article.
-
New Insights on the Evolution of Genome Content: Population Dynamics of Transposable Elements in Flies and Humans.Methods Mol Biol. 2019;1910:505-530. doi: 10.1007/978-1-4939-9074-0_16. Methods Mol Biol. 2019. PMID: 31278675 Review.
-
Horizontal transfers of transposable elements in eukaryotes: The flying genes.C R Biol. 2016 Jul-Aug;339(7-8):296-9. doi: 10.1016/j.crvi.2016.04.013. Epub 2016 May 24. C R Biol. 2016. PMID: 27234293 Review.
Cited by
-
General survey of hAT transposon superfamily with highlight on hobo element in Drosophila.Genetica. 2012 Sep;140(7-9):375-92. doi: 10.1007/s10709-012-9687-0. Epub 2012 Oct 31. Genetica. 2012. PMID: 23111927
-
How does selfing affect the dynamics of selfish transposable elements?Mob DNA. 2012 Mar 7;3:5. doi: 10.1186/1759-8753-3-5. Mob DNA. 2012. PMID: 22394388 Free PMC article.
-
The predominantly selfing plant Arabidopsis thaliana experienced a recent reduction in transposable element abundance compared to its outcrossing relative Arabidopsis lyrata.Mob DNA. 2012 Feb 7;3(1):2. doi: 10.1186/1759-8753-3-2. Mob DNA. 2012. PMID: 22313744 Free PMC article.
-
Applying mobile genetic elements for genome analysis and evolution.Mol Biotechnol. 2006 Jun;33(2):161-74. doi: 10.1385/MB:33:2:161. Mol Biotechnol. 2006. PMID: 16757803 Review.
-
Internal deletions of transposable elements: the case of Lemi elements.Genetica. 2013 Sep;141(7-9):369-79. doi: 10.1007/s10709-013-9736-3. Epub 2013 Oct 11. Genetica. 2013. PMID: 24114377
References
-
- Anxolabéhère, D., M. G. Kidwell and G. Periquet, 1988. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 5: 252–269. - PubMed
-
- Biémont, C., 1994. Dynamic equilibrium between insertion and excision of P elements in highly inbred lines from an M′ strain of Drosophila melanogaster. J. Mol. Evol. 39: 466–472. - PubMed
-
- Biémont, C., F. Lemeunier, M. P. Garcia Guerreiro, J. F. Brookfield, C. Gautier et al., 1994. Population dynamics of the copia, mdg1, mdg3, gypsy, and P transposable elements in a natural population of Drosophila melanogaster. Genet. Res. 63: 197–212. - PubMed
-
- Biémont, C., C. Nardon, G. Deceliere, D. Lepetit, C. Loevenbruck et al., 2003. Worldwide distribution of transposable element copy number in natural populations of Drosophila simulans. Evolution 57: 159–167. - PubMed
-
- Bregliano, J.-C., and M. G. Kidwell, 1983 Hybrid dysgenesis determinants, p. 363 in Mobile Genetic Elements, edited by J. A. Shapiro. Academic Press, San Diego.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

