Nuclear localization of Japanese encephalitis virus core protein enhances viral replication
- PMID: 15731239
- PMCID: PMC1075736
- DOI: 10.1128/JVI.79.6.3448-3458.2005
Nuclear localization of Japanese encephalitis virus core protein enhances viral replication
Abstract
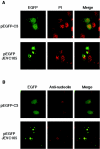
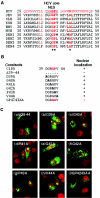
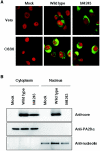
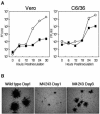
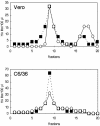
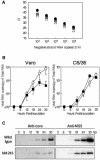
Japanese encephalitis virus (JEV) core protein was detected in both the nucleoli and cytoplasm of mammalian and insect cell lines infected with JEV or transfected with the expression plasmid of the core protein. Mutation analysis revealed that Gly(42) and Pro(43) in the core protein are essential for the nuclear and nucleolar localization. A mutant M4243 virus in which both Gly(42) and Pro(43) were replaced by Ala was recovered by plasmid-based reverse genetics. In C6/36 mosquito cells, the M4243 virus exhibited RNA replication and protein synthesis comparable to wild-type JEV, whereas propagation in Vero cells was impaired. The mutant core protein was detected in the cytoplasm but not in the nucleus of either C6/36 or Vero cell lines infected with the M4243 virus. The impaired propagation of M4243 in mammalian cells was recovered by the expression of wild-type core protein in trans but not by that of the mutant core protein. Although M4243 mutant virus exhibited a high level of neurovirulence comparable to wild-type JEV in spite of the approximately 100-fold-lower viral propagation after intracerebral inoculation to 3-week-old mice of strain Jcl:ICR, no virus was recovered from the brain after intraperitoneal inoculation of the mutant. These results indicate that nuclear localization of JEV core protein plays crucial roles not only in the replication in mammalian cells in vitro but also in the pathogenesis of encephalitis induced by JEV in vivo.
Figures

Similar articles
-
Processing of capsid protein by cathepsin L plays a crucial role in replication of Japanese encephalitis virus in neural and macrophage cells.J Virol. 2007 Aug;81(16):8477-87. doi: 10.1128/JVI.00477-07. Epub 2007 Jun 6. J Virol. 2007. PMID: 17553875 Free PMC article.
-
Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication.Microbiol Immunol. 2006;50(3):225-34. doi: 10.1111/j.1348-0421.2006.tb03789.x. Microbiol Immunol. 2006. PMID: 16547420
-
Sindbis vectors suppress secretion of subviral particles of Japanese encephalitis virus from mammalian cells infected with SIN-JEV recombinants.Virology. 1995 May 10;209(1):155-66. doi: 10.1006/viro.1995.1239. Virology. 1995. PMID: 7747465
-
The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication.Virus Res. 2003 Sep;95(1-2):13-22. doi: 10.1016/s0168-1702(03)00160-6. Virus Res. 2003. PMID: 12921992 Free PMC article. Review.
-
Molecular basis of subcellular localization of HCV core protein.Liver. 1996 Aug;16(4):221-4. doi: 10.1111/j.1600-0676.1996.tb00732.x. Liver. 1996. PMID: 8877990 Review. No abstract available.
Cited by
-
Molecular and pathological screening of the current circulation of fowlpox and pigeon pox virus in backyard birds.Poult Sci. 2024 Dec;103(12):104249. doi: 10.1016/j.psj.2024.104249. Epub 2024 Aug 22. Poult Sci. 2024. PMID: 39418793 Free PMC article.
-
Recombinant core proteins of Japanese encephalitis virus as activators of the innate immune response.Virus Genes. 2009 Feb;38(1):10-8. doi: 10.1007/s11262-008-0299-9. Epub 2008 Nov 14. Virus Genes. 2009. PMID: 19009340
-
Ribosomal stress and Tp53-mediated neuronal apoptosis in response to capsid protein of the Zika virus.Sci Rep. 2017 Nov 30;7(1):16652. doi: 10.1038/s41598-017-16952-8. Sci Rep. 2017. PMID: 29192272 Free PMC article.
-
Repeated Intravaginal Inoculation of Zika Virus Protects Cynomolgus Monkeys from Subcutaneous Superchallenge.Int J Mol Sci. 2022 Nov 13;23(22):14002. doi: 10.3390/ijms232214002. Int J Mol Sci. 2022. PMID: 36430481 Free PMC article.
-
Intracellular distribution of NS1 correlates with the infectivity and interferon antagonism of an avian influenza virus (H7N1).J Virol. 2010 Nov;84(22):11858-65. doi: 10.1128/JVI.01011-10. Epub 2010 Sep 15. J Virol. 2010. PMID: 20844052 Free PMC article.
References
-
- Aminev, A. G., S. P. Amineva, and A. C. Palmenberg. 2003. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 95:45-57. - PubMed
-
- Aminev, A. G., S. P. Amineva, and A. C. Palmenberg. 2003. Encephalomyocarditis virus (EMCV) proteins 2A and 3BCD localize to nuclei and inhibit cellular mRNA transcription but not rRNA transcription. Virus Res. 95:59-73. - PubMed
-
- Bulich, R., and J. G. Aaskov. 1992. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 73:2999-3003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

