Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction
- PMID: 15709033
- PMCID: PMC548447
- DOI: 10.1128/JVI.79.5.3139-3145.2005
Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction
Abstract
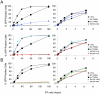
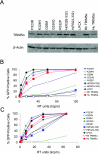
Retroviruses encounter dominant postentry restrictions in cells of particular species. Human immunodeficiency virus type 1 (HIV-1) is blocked in the cells of Old World monkeys by TRIM5alpha, a tripartite motif (TRIM) protein composed of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains. Rhesus monkey TRIM5alpha (TRIM5alpha(rh)) more potently blocks HIV-1 infection than human TRIM5alpha (TRIM5alpha(hu)). Here, by studying chimeric TRIM5alpha proteins, we demonstrate that the major determinant of anti-HIV-1 potency is the B30.2(SPRY) domain. Analysis of species-specific variation in TRIM5alpha has identified three variable regions (v1, v2, and v3) within the B30.2 domain. The TRIM5alpha proteins of Old World primates exhibit expansion, duplication, and residue variation specifically in the v1 region. Replacement of three amino acids in the N terminus of the TRIM5alpha(hu) B30.2 v1 region with the corresponding TRIM5alpha(rh) residues resulted in a TRIM5alpha molecule that restricted HIV-1 nearly as efficiently as wild-type TRIM5alpha(rh). Surprisingly, a single-amino-acid change in this region of TRIM5alpha(hu) allowed potent restriction of simian immunodeficiency virus, a phenotype not observed for either wild-type TRIM5alpha(hu) or TRIM5alpha(rh). Some of the chimeric TRIM5alpha proteins that are >98% identical to the human protein yet mediate a strong restriction of HIV-1 infection may have therapeutic utility. These observations implicate the v1 variable region of the B30.2(SPRY) domain in TRIM5alpha(rh) antiviral potency.
Figures

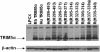


Similar articles
-
A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection.J Virol. 2005 Jul;79(14):8870-7. doi: 10.1128/JVI.79.14.8870-8877.2005. J Virol. 2005. PMID: 15994780 Free PMC article.
-
The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates.J Virol. 2005 May;79(10):6111-21. doi: 10.1128/JVI.79.10.6111-6121.2005. J Virol. 2005. PMID: 15857996 Free PMC article.
-
Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains.J Virol. 2006 Jul;80(13):6198-206. doi: 10.1128/JVI.00283-06. J Virol. 2006. PMID: 16775307 Free PMC article.
-
Retroviral restriction factors TRIM5α: therapeutic strategy to inhibit HIV-1 replication.Curr Med Chem. 2011;18(17):2649-54. doi: 10.2174/092986711795933687. Curr Med Chem. 2011. PMID: 21568899 Review.
-
Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence?Immunology. 2005 Dec;116(4):411-7. doi: 10.1111/j.1365-2567.2005.02248.x. Immunology. 2005. PMID: 16313355 Free PMC article. Review.
Cited by
-
A comparison of murine leukemia viruses that escape from human and rhesus macaque TRIM5αs.J Virol. 2013 Jun;87(11):6455-68. doi: 10.1128/JVI.03425-12. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536686 Free PMC article.
-
Duck TRIM27-L enhances MAVS signaling and is absent in chickens and turkeys.Mol Immunol. 2015 Oct;67(2 Pt B):607-15. doi: 10.1016/j.molimm.2015.07.011. Epub 2015 Aug 5. Mol Immunol. 2015. PMID: 26254985 Free PMC article.
-
Preclinical Assessment of Mutant Human TRIM5α as an Anti-HIV-1 Transgene.Hum Gene Ther. 2015 Oct;26(10):664-79. doi: 10.1089/hum.2015.059. Epub 2015 Aug 6. Hum Gene Ther. 2015. PMID: 26076730 Free PMC article.
-
Retrovirus restriction by TRIM5alpha variants from Old World and New World primates.J Virol. 2005 Apr;79(7):3930-7. doi: 10.1128/JVI.79.7.3930-3937.2005. J Virol. 2005. PMID: 15767395 Free PMC article.
-
Characterization of an amino-terminal dimerization domain from retroviral restriction factor Fv1.J Virol. 2006 Aug;80(16):8225-35. doi: 10.1128/JVI.00395-06. J Virol. 2006. PMID: 16873278 Free PMC article.
References
-
- Arts, E. J., and M. A. Wainberg. 1996. Human immunodeficiency type 1 reverse transcriptase and early events in reverse transcription. Adv. Virus Res. 46:97-163. - PubMed
-
- Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rosenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS). Science 220:868-871. - PubMed
-
- Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. - PubMed
-
- Clavel, F. 1987. HIV-2, the West African AIDS virus. AIDS 1:135-140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

