Establishment and maintenance of long-term murine gammaherpesvirus 68 latency in B cells in the absence of CD40
- PMID: 15709008
- PMCID: PMC548450
- DOI: 10.1128/JVI.79.5.2891-2899.2005
Establishment and maintenance of long-term murine gammaherpesvirus 68 latency in B cells in the absence of CD40
Abstract
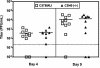
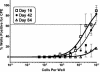
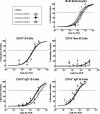
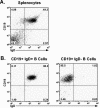
Murine gammaherpesvirus 68 (gammaHV68), like Epstein-Barr virus (EBV), establishes a chronic infection in its host by gaining access to the memory B-cell reservoir, where it persists undetected by the host's immune system. EBV encodes a membrane protein, LMP1, that appears to function as a constitutively active CD40 receptor, and is hypothesized to play a central role in EBV-driven differentiation of infected naive B cells to a memory B-cell phenotype. However, it has recently been shown that there is a critical role for CD40-CD40L interaction in B-cell immortalization by EBV (K.-I. Imadome, M. Shirakata, N. Shimizu, S. Nonoyama, and Y. Yamanashi, Proc. Natl. Acad. Sci. USA 100:7836-7840, 2003), indicating that LMP1 does not adequately recapitulate all of the necessary functions of CD40. The role of CD40 receptor expression on B cells for the establishment and maintenance of gammaHV68 latency is unclear. Data previously obtained with a competition model, demonstrated that in the face of CD40-sufficient B cells, gammaHV68 latency in CD40-deficient B cells waned over time in chimeric mice (I.-J. Kim, E. Flano, D. L. Woodland, F. E. Lund, T. D. Randall, and M. A. Blackman, J. Immunol. 171:886-892, 2003). To further investigate the role of CD40 in gammaHV68 latency in vivo, we have characterized the infection of CD40 knockout (CD40(-/-)) mice. Here we report that, consistent with previous observations, gammaHV68 efficiently established a latent infection in B cells of CD40(-/-) mice. Notably, unlike the infection of normal C57BL/6 mice, significant ex vivo reactivation from splenocytes harvested from infected CD40(-/-) mice 42 days postinfection was observed. In addition, in contrast to gammaHV68 infection of C57BL/6 mice, the frequency of infected naive B cells remained fairly stable over a 3-month period postinfection. Furthermore, a slightly higher frequency of gammaHV68 infection was observed in immunoglobulin D (IgD)-negative B cells, which was stably maintained over a period of 3 months postinfection. The presence of virus in IgD-negative B cells indicates that gammaHV68 may either directly infect memory B cells present in CD40(-/-) mice or be capable of driving differentiation of naive CD40(-/-) B cells. A possible explanation for the apparent discrepancy between the failure of gammaHV68 latency to be maintained in CD40-deficient B cells in the presence of CD40-sufficient B cells and the stable maintenance of gammaHV68 B-cell latency in CD40(-/-) mice came from examining virus replication in the lungs of infected CD40(-/-) mice, where we observed significantly higher levels of virus replication at late times postinfection compared to those in infected C57BL/6 mice. Taken together, these findings are consistent with a model in which chronic virus infection of CD40(-/-) mice is maintained through virus reactivation in the lungs and reseeding of latency reservoirs.
Figures
Similar articles
-
Role of B-cell proliferation in the establishment of gammaherpesvirus latency.J Virol. 2005 Aug;79(15):9480-91. doi: 10.1128/JVI.79.15.9480-9491.2005. J Virol. 2005. PMID: 16014911 Free PMC article.
-
Ex vivo stimulation of B cells latently infected with gammaherpesvirus 68 triggers reactivation from latency.J Virol. 2005 Apr;79(8):5227-31. doi: 10.1128/JVI.79.8.5227-5231.2005. J Virol. 2005. PMID: 15795307 Free PMC article.
-
Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency.PLoS Pathog. 2007 Jan;3(1):e11. doi: 10.1371/journal.ppat.0030011. PLoS Pathog. 2007. PMID: 17257062 Free PMC article.
-
Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner.J Virol. 2007 Jun;81(11):6134-40. doi: 10.1128/JVI.00108-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360749 Free PMC article. Review.
-
Natural history of murine gamma-herpesvirus infection.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):569-79. doi: 10.1098/rstb.2000.0779. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313012 Free PMC article. Review.
Cited by
-
Deletion of Murine Gammaherpesvirus Gene M2 in Activation-Induced Cytidine Deaminase-Expressing B Cells Impairs Host Colonization and Viral Reactivation.J Virol. 2020 Dec 9;95(1):e01933-20. doi: 10.1128/JVI.01933-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33028711 Free PMC article.
-
Conserved Gammaherpesvirus Protein Kinase Selectively Promotes Irrelevant B Cell Responses.J Virol. 2019 Apr 3;93(8):e01760-18. doi: 10.1128/JVI.01760-18. Print 2019 Apr 15. J Virol. 2019. PMID: 30728267 Free PMC article.
-
Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade.J Virol. 2008 Aug;82(15):7688-99. doi: 10.1128/JVI.02689-07. Epub 2008 May 21. J Virol. 2008. PMID: 18495760 Free PMC article.
-
RTA Occupancy of the Origin of Lytic Replication during Murine Gammaherpesvirus 68 Reactivation from B Cell Latency.Pathogens. 2017 Feb 16;6(1):9. doi: 10.3390/pathogens6010009. Pathogens. 2017. PMID: 28212352 Free PMC article.
-
Interplay of Murine Gammaherpesvirus 68 with NF-kappaB Signaling of the Host.Front Microbiol. 2016 Aug 17;7:1202. doi: 10.3389/fmicb.2016.01202. eCollection 2016. Front Microbiol. 2016. PMID: 27582728 Free PMC article. Review.
References
-
- Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. - PubMed
-
- Babcock, G. J., D. Hochberg, and A. D. Thorley-Lawson. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497-506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

