Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I
- PMID: 15708988
- PMCID: PMC548482
- DOI: 10.1128/JVI.79.5.2689-2699.2005
Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I
Abstract
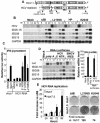
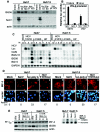
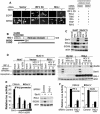
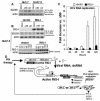
Virus-responsive signaling pathways that induce alpha/beta interferon production and engage intracellular immune defenses influence the outcome of many viral infections. The processes that trigger these defenses and their effect upon host permissiveness for specific viral pathogens are not well understood. We show that structured hepatitis C virus (HCV) genomic RNA activates interferon regulatory factor 3 (IRF3), thereby inducing interferon in cultured cells. This response is absent in cells selected for permissiveness for HCV RNA replication. Studies including genetic complementation revealed that permissiveness is due to mutational inactivation of RIG-I, an interferon-inducible cellular DExD/H box RNA helicase. Its helicase domain binds HCV RNA and transduces the activation signal for IRF3 by its caspase recruiting domain homolog. RIG-I is thus a pathogen receptor that regulates cellular permissiveness to HCV replication and, as an interferon-responsive gene, may play a key role in interferon-based therapies for the treatment of HCV infection.
Figures

Similar articles
-
Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2.Proc Natl Acad Sci U S A. 2007 Jan 9;104(2):582-7. doi: 10.1073/pnas.0606699104. Epub 2006 Dec 26. Proc Natl Acad Sci U S A. 2007. PMID: 17190814 Free PMC article.
-
DDX60L Is an Interferon-Stimulated Gene Product Restricting Hepatitis C Virus Replication in Cell Culture.J Virol. 2015 Oct;89(20):10548-68. doi: 10.1128/JVI.01297-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269178 Free PMC article.
-
Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy.Hepatology. 2007 Nov;46(5):1365-74. doi: 10.1002/hep.21829. Hepatology. 2007. PMID: 17668876
-
Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses.Semin Liver Dis. 2010 Nov;30(4):319-32. doi: 10.1055/s-0030-1267534. Epub 2010 Oct 19. Semin Liver Dis. 2010. PMID: 20960373 Review.
-
[Virus-induced expression of type I interferon genes].Uirusu. 2004 Dec;54(2):161-7. doi: 10.2222/jsv.54.161. Uirusu. 2004. PMID: 15745153 Review. Japanese.
Cited by
-
A genome-wide arrayed CRISPR screen identifies PLSCR1 as an intrinsic barrier to SARS-CoV-2 entry that recent virus variants have evolved to resist.PLoS Biol. 2024 Sep 24;22(9):e3002767. doi: 10.1371/journal.pbio.3002767. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39316623 Free PMC article.
-
Early interferon lambda production is induced by double-stranded RNA in iPS-derived hepatocyte-like cells.Oxf Open Immunol. 2024 Jun 6;5(1):iqae004. doi: 10.1093/oxfimm/iqae004. eCollection 2024. Oxf Open Immunol. 2024. PMID: 39193476 Free PMC article.
-
Proofreading mechanisms of the innate immune receptor RIG-I: distinguishing self and viral RNA.Biochem Soc Trans. 2024 Jun 26;52(3):1131-1148. doi: 10.1042/BST20230724. Biochem Soc Trans. 2024. PMID: 38884803 Free PMC article. Review.
-
The NS1 protein of influenza B virus binds 5'-triphosphorylated dsRNA to suppress RIG-I activation and the host antiviral response.bioRxiv [Preprint]. 2024 Jan 24:2023.09.25.559316. doi: 10.1101/2023.09.25.559316. bioRxiv. 2024. PMID: 38328244 Free PMC article. Preprint.
-
Unraveling the dynamics of hepatitis C virus adaptive mutations and their impact on antiviral responses in primary human hepatocytes.J Virol. 2024 Mar 19;98(3):e0192123. doi: 10.1128/jvi.01921-23. Epub 2024 Feb 6. J Virol. 2024. PMID: 38319104 Free PMC article.
References
-
- Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antivir. Res. 52:1-17. - PubMed
-
- Edelmann, K. H., S. Richardson-Burns, L. Alexopoulou, K. L. Tyler, R. A. Flavell, and M. B. Oldstone. 2004. Does Toll-like receptor 3 play a biological role in virus infections? Virology 322:231-238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

