The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p
- PMID: 15706350
- PMCID: PMC554118
- DOI: 10.1038/sj.emboj.7600560
The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p
Abstract
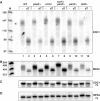
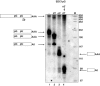
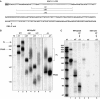
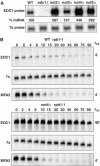
A major mechanism of eukaryotic mRNA degradation initiates with deadenylation followed by decapping and 5' to 3' degradation. We demonstrate that the yeast EDC1 mRNA, which encodes a protein that enhances decapping, has unique properties and is both protected from deadenylation and undergoes deadenylation-independent decapping. The 3' UTR of the EDC1 mRNA is sufficient for both protection from deadenylation and deadenylation-independent decapping and an extended poly(U) tract within the 3' UTR is required. These observations highlight the diverse forms of decapping regulation and identify a feedback loop that can compensate for decreases in activity of the decapping enzyme. Surprisingly, the decapping of the EDC1 mRNA is slowed by the loss of Not2p, Not4p, and Not5p, which interact with the Ccr4p/Pop2p deadenylase complex. This indicates that the Not proteins can affect decapping, which suggests a possible link between the mRNA deadenylation and decapping machinery.
Figures

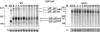

Similar articles
-
The deadenylase components Not2p, Not3p, and Not5p promote mRNA decapping.RNA. 2016 May;22(5):709-21. doi: 10.1261/rna.054742.115. Epub 2016 Mar 7. RNA. 2016. PMID: 26952104 Free PMC article.
-
Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae.EMBO J. 2002 Mar 15;21(6):1427-36. doi: 10.1093/emboj/21.6.1427. EMBO J. 2002. PMID: 11889048 Free PMC article.
-
The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation.Nucleic Acids Res. 2001 Jun 15;29(12):2448-55. doi: 10.1093/nar/29.12.2448. Nucleic Acids Res. 2001. PMID: 11410650 Free PMC article.
-
Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
-
Eukaryotic mRNA decapping factors: molecular mechanisms and activity.FEBS J. 2023 Nov;290(21):5057-5085. doi: 10.1111/febs.16626. Epub 2022 Sep 30. FEBS J. 2023. PMID: 36098474 Free PMC article. Review.
Cited by
-
Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies.Mol Cell Biol. 2010 Sep;30(17):4308-23. doi: 10.1128/MCB.00429-10. Epub 2010 Jun 28. Mol Cell Biol. 2010. PMID: 20584987 Free PMC article.
-
The RNA helicase Dhh1p cooperates with Rbp1p to promote porin mRNA decay via its non-conserved C-terminal domain.Nucleic Acids Res. 2012 Feb;40(3):1331-44. doi: 10.1093/nar/gkr803. Epub 2011 Oct 13. Nucleic Acids Res. 2012. PMID: 21998293 Free PMC article.
-
The control of mRNA decapping and P-body formation.Mol Cell. 2008 Dec 5;32(5):605-15. doi: 10.1016/j.molcel.2008.11.001. Mol Cell. 2008. PMID: 19061636 Free PMC article. Review.
-
The deadenylase components Not2p, Not3p, and Not5p promote mRNA decapping.RNA. 2016 May;22(5):709-21. doi: 10.1261/rna.054742.115. Epub 2016 Mar 7. RNA. 2016. PMID: 26952104 Free PMC article.
-
Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Jan;27(1):92-101. doi: 10.1128/MCB.01023-06. Epub 2006 Oct 30. Mol Cell Biol. 2007. PMID: 17074811 Free PMC article.
References
-
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A (2004) Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell 15: 5–15 - PubMed
-
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

