Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae
- PMID: 15705057
- PMCID: PMC1138978
- DOI: 10.1042/BJ20050160
Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae
Abstract
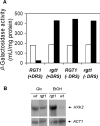
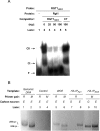
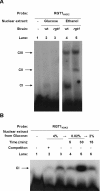
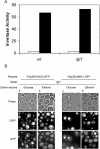
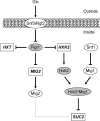
Expression of HXK2, a gene encoding a Saccharomyces cerevisiae bifunctional protein with catalytic and regulatory functions, is controlled by glucose availability, being activated in the presence of glucose and inhibited when the levels of the sugar are low. In the present study, we identified Rgt1 as a transcription factor that, together with the Med8 protein, is essential for repression of the HXK2 gene in the absence of glucose. Rgt1 represses HXK2 expression by binding specifically to the motif (CGGAAAA) located at -395 bp relative to the ATG translation start codon in the HXK2 promoter. Disruption of the RGT1 gene causes an 18-fold increase in the level of HXK2 transcript in the absence of glucose. Rgt1 binds to the RGT1 element of HXK2 promoter in a glucose-dependent manner, and the repression of target gene depends on binding of Rgt1 to DNA. The physiological significance of the connection between two glucose-signalling pathways, the Snf3/Rgt2 that causes glucose induction and the Mig1/Hxk2 that causes glucose repression, was also analysed.
Figures
Similar articles
-
Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter.Nucleic Acids Res. 2006 Mar 9;34(5):1427-38. doi: 10.1093/nar/gkl028. Print 2006. Nucleic Acids Res. 2006. PMID: 16528100 Free PMC article.
-
How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose.Genetics. 2005 Feb;169(2):583-94. doi: 10.1534/genetics.104.034512. Epub 2004 Oct 16. Genetics. 2005. PMID: 15489524 Free PMC article.
-
DNA-binding properties of the yeast Rgt1 repressor.Biochimie. 2009 Feb;91(2):300-3. doi: 10.1016/j.biochi.2008.09.002. Epub 2008 Oct 7. Biochimie. 2009. PMID: 18950675 Free PMC article.
-
The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae.FEMS Microbiol Rev. 2002 Mar;26(1):83-90. doi: 10.1111/j.1574-6976.2002.tb00600.x. FEMS Microbiol Rev. 2002. PMID: 12007644 Review.
-
Glucose repression in the yeast Saccharomyces cerevisiae.Mol Microbiol. 1992 Jan;6(1):15-21. doi: 10.1111/j.1365-2958.1992.tb00832.x. Mol Microbiol. 1992. PMID: 1310793 Review.
Cited by
-
Moonlighting proteins in yeasts.Microbiol Mol Biol Rev. 2008 Mar;72(1):197-210, table of contents. doi: 10.1128/MMBR.00036-07. Microbiol Mol Biol Rev. 2008. PMID: 18322039 Free PMC article. Review.
-
A fast method to distinguish between fermentative and respiratory metabolisms in single yeast cells.iScience. 2023 Dec 21;27(1):108767. doi: 10.1016/j.isci.2023.108767. eCollection 2024 Jan 19. iScience. 2023. PMID: 38235328 Free PMC article.
-
Genomic expression patterns in cell separation mutants of Schizosaccharomyces pombe defective in the genes sep10 ( + ) and sep15 ( + ) coding for the Mediator subunits Med31 and Med8.Mol Genet Genomics. 2008 Mar;279(3):225-38. doi: 10.1007/s00438-007-0296-z. Epub 2007 Oct 7. Mol Genet Genomics. 2008. PMID: 17922236
-
PKA regulatory subunit Bcy1 couples growth, lipid metabolism, and fermentation during anaerobic xylose growth in Saccharomyces cerevisiae.PLoS Genet. 2023 Jul 6;19(7):e1010593. doi: 10.1371/journal.pgen.1010593. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37410771 Free PMC article.
-
Connection between the Rag4 glucose sensor and the KlRgt1 repressor in Kluyveromyces lactis.Genetics. 2006 Oct;174(2):617-26. doi: 10.1534/genetics.106.059766. Epub 2006 Jun 18. Genetics. 2006. PMID: 16783006 Free PMC article.
References
-
- Carlson M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999;2:202–207. - PubMed
-
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. - PubMed
-
- Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J. M. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 2004;29:556–564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

