Catalytic mechanism of Escherichia coli ribonuclease III: kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis
- PMID: 15699182
- PMCID: PMC549391
- DOI: 10.1093/nar/gki197
Catalytic mechanism of Escherichia coli ribonuclease III: kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis
Abstract
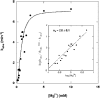
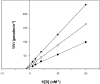
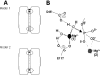
Escherichia coli ribonuclease III (RNase III; EC 3.1.24) is a double-stranded(ds)-RNA-specific endonuclease with key roles in diverse RNA maturation and decay pathways. E.coli RNase III is a member of a structurally distinct superfamily that includes Dicer, a central enzyme in the mechanism of RNA interference. E.coli RNase III requires a divalent metal ion for activity, with Mg2+ as the preferred species. However, neither the function(s) nor the number of metal ions involved in catalysis is known. To gain information on metal ion involvement in catalysis, the rate of cleavage of the model substrate R1.1 RNA was determined as a function of Mg2+ concentration. Single-turnover conditions were applied, wherein phosphodiester cleavage was the rate-limiting event. The measured Hill coefficient (n (H)) is 2.0 +/- 0.1, indicative of the involvement of two Mg2+ ions in phosphodiester hydrolysis. It is also shown that 2-hydroxy-4H-isoquinoline-1,3-dione--an inhibitor of ribonucleases that employ two divalent metal ions in their catalytic sites--inhibits E.coli RNase III cleavage of R1.1 RNA. The IC50 for the compound is 14 microM for the Mg2+-supported reaction, and 8 microM for the Mn2+-supported reaction. The compound exhibits noncompetitive inhibitory kinetics, indicating that it does not perturb substrate binding. Neither the O-methylated version of the compound nor the unsubstituted imide inhibit substrate cleavage, which is consistent with a specific interaction of the N-hydroxyimide with two closely positioned divalent metal ions. A preliminary model is presented for functional roles of two divalent metal ions in the RNase III catalytic mechanism.
Figures

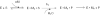
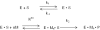
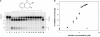
Similar articles
-
Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant.Biochemistry. 2001 Apr 24;40(16):5102-10. doi: 10.1021/bi010022d. Biochemistry. 2001. PMID: 11305928
-
Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis.J Mol Biol. 1999 Jul 9;290(2):433-45. doi: 10.1006/jmbi.1999.2890. J Mol Biol. 1999. PMID: 10390342
-
Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro.Biochem J. 2008 Feb 15;410(1):39-48. doi: 10.1042/BJ20071047. Biochem J. 2008. PMID: 17953512
-
Ribonuclease III mechanisms of double-stranded RNA cleavage.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):31-48. doi: 10.1002/wrna.1195. Epub 2013 Sep 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24124076 Free PMC article. Review.
-
Ribonuclease revisited: structural insights into ribonuclease III family enzymes.Curr Opin Struct Biol. 2007 Feb;17(1):138-45. doi: 10.1016/j.sbi.2006.12.002. Epub 2006 Dec 27. Curr Opin Struct Biol. 2007. PMID: 17194582 Review.
Cited by
-
2-(Benz-yloxy)iso-quinoline-1,3(2H,4H)-dione.Acta Crystallogr Sect E Struct Rep Online. 2013 Jul 24;69(Pt 8):o1311. doi: 10.1107/S1600536813019934. eCollection 2013. Acta Crystallogr Sect E Struct Rep Online. 2013. PMID: 24109385 Free PMC article.
-
Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.Mol Cell. 2022 Nov 3;82(21):4049-4063.e6. doi: 10.1016/j.molcel.2022.09.002. Epub 2022 Sep 30. Mol Cell. 2022. PMID: 36182693 Free PMC article.
-
Global control of cellular physiology by biomolecular condensates through modulation of electrochemical equilibria.bioRxiv [Preprint]. 2023 Oct 20:2023.10.19.563018. doi: 10.1101/2023.10.19.563018. bioRxiv. 2023. PMID: 37904914 Free PMC article. Preprint.
-
Crystal structure and putative function of small Toprim domain-containing protein from Bacillus stearothermophilus.Proteins. 2008 Feb 1;70(2):311-9. doi: 10.1002/prot.21511. Proteins. 2008. PMID: 17705269 Free PMC article.
-
Mechanism of Ribonuclease III Catalytic Regulation by Serine Phosphorylation.Sci Rep. 2016 May 6;6:25448. doi: 10.1038/srep25448. Sci Rep. 2016. PMID: 27150669 Free PMC article.
References
-
- Court D.L. RNA processing and degradation by RNase III. In: Belasco J.G., Brawerman G., editors. Control of Messenger RNA Stability. New York: Academic Press; 1993. pp. 71–116.
-
- LaMontagne B., Larose S., Boulanger J., AbouElela S. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 2001;3:71–78. - PubMed
-
- Nicholson A.W. The ribonuclease superfamily: forms and functions in RNA maturation, decay, and gene silencing. In: Hannon G.J., editor. RNAi: A Guide to Gene Silencing. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. pp. 149–174.
-
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate nuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

