Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export
- PMID: 15692572
- PMCID: PMC549612
- DOI: 10.1038/sj.emboj.7600527
Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export
Abstract
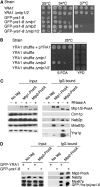
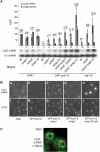
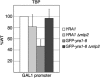
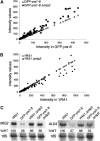
The mRNA export adaptor Yra1p/REF contributes to nascent mRNP assembly and recruitment of the export receptor Mex67p. yra1 mutants exhibit mRNA export defects and a decrease in LacZ reporter and certain endogenous transcripts. The loss of Mlp1p/Mlp2p, two TPR-like proteins attached to nuclear pores, rescues LacZ mRNA levels and increases their appearance in the cytoplasm, without restoring bulk poly(A)+ RNA export. Chromatin immunoprecipitation, FISH and pulse-chase experiments indicate that Mlps downregulate LacZ mRNA synthesis in a yra1 mutant strain. Microarray analyses reveal that Mlp2p also reduces a subset of cellular transcripts in the yra1 mutant. Finally, we show that Yra1p genetically interacts with the shuttling mRNA-binding protein Nab2p and that loss of Mlps rescues the growth defect of yra1 and nab2 but not other mRNA export mutants. We propose that Nab2p and Yra1p are required for proper mRNP docking to the Mlp platform. Defects in Yra1p prevent mRNPs from crossing the Mlp gate and this block negatively feeds back on the transcription of a subset of genes, suggesting that Mlps link mRNA transcription and export.
Figures


Similar articles
-
The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p.Mol Cell Biol. 2001 Jul;21(13):4219-32. doi: 10.1128/MCB.21.13.4219-4232.2001. Mol Cell Biol. 2001. PMID: 11390651 Free PMC article.
-
Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export.Genes Dev. 2010 Sep 1;24(17):1927-38. doi: 10.1101/gad.583310. Genes Dev. 2010. PMID: 20810649 Free PMC article.
-
YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA.Mol Cell. 2007 Feb 23;25(4):559-73. doi: 10.1016/j.molcel.2007.01.012. Mol Cell. 2007. PMID: 17317628 Free PMC article.
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair.Curr Genet. 2020 Feb;66(1):63-71. doi: 10.1007/s00294-019-01011-8. Epub 2019 Jul 10. Curr Genet. 2020. PMID: 31292684 Review.
-
[Coordinated pathway of nuclear mRNA export reveled by the genetic analysis in yeast].Tanpakushitsu Kakusan Koso. 2009 Dec;54(16 Suppl):2102-8. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089625 Review. Japanese. No abstract available.
Cited by
-
Multiple crosstalks between mRNA biogenesis and SUMO.Chromosoma. 2013 Oct;122(5):387-99. doi: 10.1007/s00412-013-0408-y. Epub 2013 Apr 14. Chromosoma. 2013. PMID: 23584125
-
Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination.Microb Cell. 2024 Feb 23;11:69-78. doi: 10.15698/mic2024.02.816. eCollection 2024. Microb Cell. 2024. PMID: 38414808 Free PMC article.
-
Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes.Mol Cell Biol. 2006 Nov;26(21):7858-70. doi: 10.1128/MCB.00870-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954382 Free PMC article.
-
Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes.PLoS One. 2010 Jul 12;5(7):e11540. doi: 10.1371/journal.pone.0011540. PLoS One. 2010. PMID: 20634951 Free PMC article.
-
Nuclear mRNA Export and Aging.Int J Mol Sci. 2022 May 13;23(10):5451. doi: 10.3390/ijms23105451. Int J Mol Sci. 2022. PMID: 35628261 Free PMC article. Review.
References
-
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 - PubMed
-
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 - PubMed
-
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

