Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins
- PMID: 15692568
- PMCID: PMC548658
- DOI: 10.1038/sj.emboj.7600553
Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins
Abstract
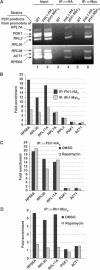
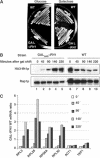
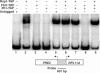
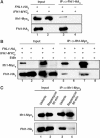
The 138 genes encoding the 79 ribosomal proteins (RPs) of Saccharomyces cerevisiae form the tightest cluster of coordinately regulated genes in nearly all transcriptome experiments. The basis for this observation remains unknown. We now provide evidence that two factors, Fhl1p and Ifh1p, are key players in the transcription of RP genes. Both are found at transcribing RP genes in vivo. Ifh1p, but not Fhl1p, leaves the RP genes when transcription is repressed. The occupancy of the RP genes by Ifh1p depends on its interaction with the phospho-peptide recognizing forkhead-associated domain of Fhl1p. Disruption of this interaction is severely deleterious to ribosome synthesis and cell growth. Loss of functional Fhl1p leads to cells that have only 20% the normal amount of RNA and that synthesize ribosomes at only 5-10% the normal rate. Homeostatic mechanisms within the cell respond by reducing the transcription of rRNA to match the output of RPs, and by reducing the global transcription of mRNA to match the capacity of the translational apparatus.
Figures


Similar articles
-
The yeast CPC2/ASC1 gene is regulated by the transcription factors Fhl1p and Ifh1p.Curr Genet. 2006 Apr;49(4):218-28. doi: 10.1007/s00294-005-0049-7. Epub 2006 Jan 10. Curr Genet. 2006. PMID: 16402205
-
The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes.Nature. 2004 Dec 23;432(7020):1054-8. doi: 10.1038/nature03175. Nature. 2004. PMID: 15616568
-
Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1.Nature. 2004 Dec 23;432(7020):1058-61. doi: 10.1038/nature03200. Nature. 2004. PMID: 15616569
-
DNA protein interactions at the rRNA of Saccharomyces cerevisiae.Ital J Biochem. 2007 Jun;56(2):81-90. Ital J Biochem. 2007. PMID: 17722648 Review.
-
Synthesis of ribosomes in Saccharomyces cerevisiae.Microbiol Rev. 1989 Jun;53(2):256-71. doi: 10.1128/mr.53.2.256-271.1989. Microbiol Rev. 1989. PMID: 2666845 Free PMC article. Review.
Cited by
-
Mechanisms coordinating ribosomal protein gene transcription in response to stress.Nucleic Acids Res. 2020 Nov 18;48(20):11408-11420. doi: 10.1093/nar/gkaa852. Nucleic Acids Res. 2020. PMID: 33084907 Free PMC article.
-
Interaction of TOR and PKA Signaling in S. cerevisiae.Biomolecules. 2022 Jan 26;12(2):210. doi: 10.3390/biom12020210. Biomolecules. 2022. PMID: 35204711 Free PMC article. Review.
-
Precise regulation of gene expression dynamics favors complex promoter architectures.PLoS Comput Biol. 2009 Jan;5(1):e1000279. doi: 10.1371/journal.pcbi.1000279. Epub 2009 Jan 30. PLoS Comput Biol. 2009. PMID: 19180182 Free PMC article.
-
Evolution of Robustness to Protein Mistranslation by Accelerated Protein Turnover.PLoS Biol. 2015 Nov 6;13(11):e1002291. doi: 10.1371/journal.pbio.1002291. eCollection 2015. PLoS Biol. 2015. PMID: 26544557 Free PMC article.
-
Expression of FHL1 in gastric cancer tissue and its correlation with the invasion and metastasis of gastric cancer.Mol Cell Biochem. 2012 Apr;363(1-2):93-9. doi: 10.1007/s11010-011-1161-2. Epub 2011 Dec 6. Mol Cell Biochem. 2012. PMID: 22143536
References
-
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR (2001) A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell 7: 741–751 - PubMed
-
- Beer MA, Tavazoie S (2004) Predicting gene expression from sequence. Cell 117: 185–198 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

