Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3
- PMID: 15681410
- PMCID: PMC546554
- DOI: 10.1128/JVI.79.4.2079-2086.2005
Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3
Abstract
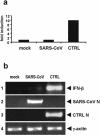
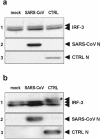
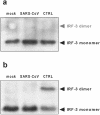
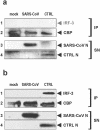
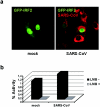
Severe acute respiratory syndrome (SARS) is caused by a novel coronavirus termed SARS-CoV. We and others have previously shown that the replication of SARS-CoV can be suppressed by exogenously added interferon (IFN), a cytokine which is normally synthesized by cells as a reaction to virus infection. Here, we demonstrate that SARS-CoV escapes IFN-mediated growth inhibition by preventing the induction of IFN-beta. In SARS-CoV-infected cells, no endogenous IFN-beta transcripts and no IFN-beta promoter activity were detected. Nevertheless, the transcription factor interferon regulatory factor 3 (IRF-3), which is essential for IFN-beta promoter activity, was transported from the cytoplasm to the nucleus early after infection with SARS-CoV. However, at a later time point in infection, IRF-3 was again localized in the cytoplasm. By contrast, IRF-3 remained in the nucleus of cells infected with the IFN-inducing control virus Bunyamwera delNSs. Other signs of IRF-3 activation such as hyperphosphorylation, homodimer formation, and recruitment of the coactivator CREB-binding protein (CBP) were found late after infection with the control virus but not with SARS-CoV. Our data suggest that nuclear transport of IRF-3 is an immediate-early reaction to virus infection and may precede its hyperphosphorylation, homodimer formation, and binding to CBP. In order to escape activation of the IFN system, SARS-CoV appears to block a step after the early nuclear transport of IRF-3.
Figures


Similar articles
-
The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3.J Virol. 2003 Jul;77(14):7945-56. doi: 10.1128/jvi.77.14.7945-7956.2003. J Virol. 2003. PMID: 12829834 Free PMC article.
-
Interferon priming enables cells to partially overturn the SARS coronavirus-induced block in innate immune activation.J Gen Virol. 2009 Nov;90(Pt 11):2686-2694. doi: 10.1099/vir.0.013599-0. Epub 2009 Jul 22. J Gen Virol. 2009. PMID: 19625461 Free PMC article.
-
Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-κB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP.J Virol. 2013 Sep;87(17):9788-801. doi: 10.1128/JVI.01440-13. Epub 2013 Jul 3. J Virol. 2013. PMID: 23824799 Free PMC article.
-
Triggering the interferon response: the role of IRF-3 transcription factor.J Interferon Cytokine Res. 1999 Jan;19(1):1-13. doi: 10.1089/107999099314360. J Interferon Cytokine Res. 1999. PMID: 10048763 Review.
-
On the role of IRF in host defense.J Interferon Cytokine Res. 2002 Jan;22(1):59-71. doi: 10.1089/107999002753452665. J Interferon Cytokine Res. 2002. PMID: 11846976 Review.
Cited by
-
A Review of Antibiotic Efficacy in COVID-19 Control.J Immunol Res. 2023 Oct 10;2023:6687437. doi: 10.1155/2023/6687437. eCollection 2023. J Immunol Res. 2023. PMID: 37854054 Free PMC article. Review.
-
Differential Type-I Interferon Response in Buffy Coat Transcriptome of Individuals Infected with SARS-CoV-2 Gamma and Delta Variants.Int J Mol Sci. 2023 Aug 24;24(17):13146. doi: 10.3390/ijms241713146. Int J Mol Sci. 2023. PMID: 37685953 Free PMC article.
-
Add fuel to the fire: Inflammation and immune response in lung cancer combined with COVID-19.Front Immunol. 2023 Mar 23;14:1174184. doi: 10.3389/fimmu.2023.1174184. eCollection 2023. Front Immunol. 2023. PMID: 37033918 Free PMC article. Review.
-
The role of dendritic cells in COVID-19 infection.Emerg Microbes Infect. 2023 Dec;12(1):2195019. doi: 10.1080/22221751.2023.2195019. Emerg Microbes Infect. 2023. PMID: 36946172 Free PMC article. Review.
-
Comparative Upper Respiratory Tract Transcriptomic Profiling Reveals a Potential Role of Early Activation of Interferon Pathway in Severe COVID-19.Viruses. 2022 Oct 1;14(10):2182. doi: 10.3390/v14102182. Viruses. 2022. PMID: 36298737 Free PMC article.
References
-
- Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. - PubMed
-
- Baudoux, P., L. Besnardeau, C. Carrat, P. Rottier, B. Charley, and H. Laude. 1998. Interferon alpha inducing property of coronavirus particles and pseudoparticles. Adv. Exp. Med. Biol. 440:377-386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

