Avian influenza H5N1 in tigers and leopards
- PMID: 15663858
- PMCID: PMC3323383
- DOI: 10.3201/eid1012.040759
Avian influenza H5N1 in tigers and leopards
Abstract
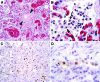
Influenza virus is not known to affect wild felids. We demonstrate that avian influenza A (H5N1) virus caused severe pneumonia in tigers and leopards that fed on infected poultry carcasses. This finding extends the host range of influenza virus and has implications for influenza virus epidemiology and wildlife conservation.
Figures

Similar articles
-
Probable tiger-to-tiger transmission of avian influenza H5N1.Emerg Infect Dis. 2005 May;11(5):699-701. doi: 10.3201/eid1105.050007. Emerg Infect Dis. 2005. PMID: 15890122 Free PMC article.
-
Genetic characterization of H5N1 influenza A viruses isolated from zoo tigers in Thailand.Virology. 2006 Jan 20;344(2):480-91. doi: 10.1016/j.virol.2005.08.032. Epub 2005 Sep 27. Virology. 2006. PMID: 16194557
-
Fatal influenza A (H5N1) virus Infection in zoo-housed Tigers in Yunnan Province, China.Sci Rep. 2016 May 10;6:25845. doi: 10.1038/srep25845. Sci Rep. 2016. PMID: 27162026 Free PMC article.
-
Influenza virus infections in dogs and cats.Vet Immunol Immunopathol. 2010 Mar 15;134(1-2):54-60. doi: 10.1016/j.vetimm.2009.10.009. Epub 2009 Oct 14. Vet Immunol Immunopathol. 2010. PMID: 19896216 Review.
-
An overview of the epidemic of highly pathogenic H5N1 avian influenza virus in Egypt: epidemiology and control challenges.Epidemiol Infect. 2011 May;139(5):647-57. doi: 10.1017/S0950268810003122. Epub 2011 Feb 1. Epidemiol Infect. 2011. PMID: 21281550 Review.
Cited by
-
Susceptibility to and transmission of H5N1 and H7N1 highly pathogenic avian influenza viruses in bank voles (Myodes glareolus).Vet Res. 2015 May 13;46(1):51. doi: 10.1186/s13567-015-0184-1. Vet Res. 2015. PMID: 25963535 Free PMC article.
-
Avian influenza outbreaks in domestic cats: another reason to consider slaughter-free cell-cultured poultry?Front Microbiol. 2023 Dec 15;14:1283361. doi: 10.3389/fmicb.2023.1283361. eCollection 2023. Front Microbiol. 2023. PMID: 38163084 Free PMC article. Review.
-
Highly pathogenic avian influenza virus H7N7 isolated from a fatal human case causes respiratory disease in cats but does not spread systemically.Am J Pathol. 2010 Nov;177(5):2185-90. doi: 10.2353/ajpath.2010.100401. Epub 2010 Sep 16. Am J Pathol. 2010. PMID: 20847292 Free PMC article.
-
Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals.J Virol. 2005 Sep;79(18):11788-800. doi: 10.1128/JVI.79.18.11788-11800.2005. J Virol. 2005. PMID: 16140756 Free PMC article.
-
Comparative pathogenesis of an avian H5N2 and a swine H1N1 influenza virus in pigs.PLoS One. 2009 Aug 17;4(8):e6662. doi: 10.1371/journal.pone.0006662. PLoS One. 2009. PMID: 19684857 Free PMC article.
References
-
- Cases of influenza A (H5N1)—Thailand, 2004. MMWR Morb Mortal Wkly Rep. 2004;53:100–3. - PubMed
-
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2). Cladistics. 1989;5:164–6.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
