Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome
- PMID: 15655079
- PMCID: PMC546205
- DOI: 10.1186/1471-2172-6-2
Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome
Abstract
Background: Severe acute respiratory syndrome (SARS) emerged in later February 2003, as a new epidemic form of life-threatening infection caused by a novel coronavirus. However, the immune-pathogenesis of SARS is poorly understood. To understand the host response to this pathogen, we investigated the gene expression profiles of peripheral blood mononuclear cells (PBMCs) derived from SARS patients, and compared with healthy controls.
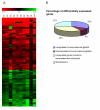
Results: The number of differentially expressed genes was found to be 186 under stringent filtering criteria of microarray data analysis. Several genes were highly up-regulated in patients with SARS, such as, the genes coding for Lactoferrin, S100A9 and Lipocalin 2. The real-time PCR method verified the results of the gene array analysis and showed that those genes that were up-regulated as determined by microarray analysis were also found to be comparatively up-regulated by real-time PCR analysis.
Conclusions: This differential gene expression profiling of PBMCs from patients with SARS strongly suggests that the response of SARS affected patients seems to be mainly an innate inflammatory response, rather than a specific immune response against a viral infection, as we observed a complete lack of cytokine genes usually triggered during a viral infection. Our study shows for the first time how the immune system responds to the SARS infection, and opens new possibilities for designing new diagnostics and treatments for this new life-threatening disease.
Figures
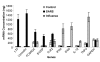
Similar articles
-
Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome.J Virol. 2007 Aug;81(16):8692-706. doi: 10.1128/JVI.00527-07. Epub 2007 May 30. J Virol. 2007. PMID: 17537853 Free PMC article.
-
[Pathologic study of circulating blood leukocytes in severe acute respiratory syndrome].Zhonghua Yi Xue Za Zhi. 2003 Dec 25;83(24):2137-41. Zhonghua Yi Xue Za Zhi. 2003. PMID: 14720422 Chinese.
-
Gene expression profiles in peripheral blood mononuclear cells of SARS patients.World J Gastroenterol. 2005 Aug 28;11(32):5037-43. doi: 10.3748/wjg.v11.i32.5037. World J Gastroenterol. 2005. PMID: 16124062 Free PMC article.
-
Differential gene expression in drug hypersensitivity reactions: induction of alarmins in severe bullous diseases.Br J Dermatol. 2010 May;162(5):1014-22. doi: 10.1111/j.1365-2133.2009.09627.x. Epub 2010 Feb 25. Br J Dermatol. 2010. PMID: 20030638
-
Mannan oligosaccharide modulates gene expression profile in pigs experimentally infected with porcine reproductive and respiratory syndrome virus.J Anim Sci. 2011 Oct;89(10):3016-29. doi: 10.2527/jas.2010-3366. Epub 2011 May 27. J Anim Sci. 2011. PMID: 21622880 Clinical Trial.
Cited by
-
An integrative drug repositioning framework discovered a potential therapeutic agent targeting COVID-19.Signal Transduct Target Ther. 2021 Apr 24;6(1):165. doi: 10.1038/s41392-021-00568-6. Signal Transduct Target Ther. 2021. PMID: 33895786 Free PMC article.
-
Nasal disinfection for the prevention and control of COVID-19: A scoping review on potential chemo-preventive agents.Int J Hyg Environ Health. 2020 Sep;230:113605. doi: 10.1016/j.ijheh.2020.113605. Epub 2020 Aug 18. Int J Hyg Environ Health. 2020. PMID: 32898838 Free PMC article.
-
Unraveling the complexities of the interferon response during SARS-CoV infection.Future Virol. 2009 Jan 1;4(1):71-78. doi: 10.2217/17460794.4.1.71. Future Virol. 2009. PMID: 19885368 Free PMC article.
-
Thrombosis-related circulating miR-16-5p is associated with disease severity in patients hospitalised for COVID-19.RNA Biol. 2022 Jan;19(1):963-979. doi: 10.1080/15476286.2022.2100629. RNA Biol. 2022. PMID: 35938548 Free PMC article.
-
Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans.PLoS One. 2011;6(8):e23710. doi: 10.1371/journal.pone.0023710. Epub 2011 Aug 22. PLoS One. 2011. PMID: 21887302 Free PMC article.
References
-
- Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet . 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. - DOI - PMC - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med . 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

