Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event
- PMID: 15650185
- PMCID: PMC544107
- DOI: 10.1128/JVI.79.3.1595-1604.2005
Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event
Abstract
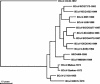
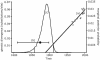
Coronaviruses are enveloped, positive-stranded RNA viruses with a genome of approximately 30 kb. Based on genetic similarities, coronaviruses are classified into three groups. Two group 2 coronaviruses, human coronavirus OC43 (HCoV-OC43) and bovine coronavirus (BCoV), show remarkable antigenic and genetic similarities. In this study, we report the first complete genome sequence (30,738 nucleotides) of the prototype HCoV-OC43 strain (ATCC VR759). Complete genome and open reading frame (ORF) analyses were performed in comparison to the BCoV genome. In the region between the spike and membrane protein genes, a 290-nucleotide deletion is present, corresponding to the absence of BCoV ORFs ns4.9 and ns4.8. Nucleotide and amino acid similarity percentages were determined for the major HCoV-OC43 ORFs and for those of other group 2 coronaviruses. The highest degree of similarity is demonstrated between HCoV-OC43 and BCoV in all ORFs with the exception of the E gene. Molecular clock analysis of the spike gene sequences of BCoV and HCoV-OC43 suggests a relatively recent zoonotic transmission event and dates their most recent common ancestor to around 1890. An evolutionary rate in the order of 4 x 10(-4) nucleotide changes per site per year was estimated. This is the first animal-human zoonotic pair of coronaviruses that can be analyzed in order to gain insights into the processes of adaptation of a nonhuman coronavirus to a human host, which is important for understanding the interspecies transmission events that led to the origin of the severe acute respiratory syndrome outbreak.
Figures

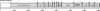


Similar articles
-
Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43.J Virol. 2006 Jul;80(14):7270-4. doi: 10.1128/JVI.02675-05. J Virol. 2006. PMID: 16809333 Free PMC article.
-
Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains.J Gen Virol. 2013 Sep;94(Pt 9):2036-2049. doi: 10.1099/vir.0.054940-0. Epub 2013 Jun 26. J Gen Virol. 2013. PMID: 23804565
-
Human respiratory coronavirus OC43: genetic stability and neuroinvasion.J Virol. 2004 Aug;78(16):8824-34. doi: 10.1128/JVI.78.16.8824-8834.2004. J Virol. 2004. PMID: 15280490 Free PMC article.
-
Hosts and Sources of Endemic Human Coronaviruses.Adv Virus Res. 2018;100:163-188. doi: 10.1016/bs.aivir.2018.01.001. Epub 2018 Feb 16. Adv Virus Res. 2018. PMID: 29551135 Free PMC article. Review.
-
Identification of new human coronaviruses.Expert Rev Anti Infect Ther. 2007 Apr;5(2):245-53. doi: 10.1586/14787210.5.2.245. Expert Rev Anti Infect Ther. 2007. PMID: 17402839 Review.
Cited by
-
First isolation of bovine coronavirus with a three-amino-acid deletion in the N gene causing severe respiratory and digestive disease in calve.Front Microbiol. 2024 Oct 1;15:1466096. doi: 10.3389/fmicb.2024.1466096. eCollection 2024. Front Microbiol. 2024. PMID: 39411436 Free PMC article.
-
Persistence of two coronaviruses and efficacy of steam vapor disinfection on two types of carpet.Virol J. 2024 Sep 2;21(1):207. doi: 10.1186/s12985-024-02478-9. Virol J. 2024. PMID: 39223556 Free PMC article.
-
Diversifying selection identified in immune epitopes of bovine coronavirus isolates from Irish cattle.J Gen Virol. 2024 Aug;105(8):002019. doi: 10.1099/jgv.0.002019. J Gen Virol. 2024. PMID: 39158563
-
Genetic characterization of bovine coronavirus strain isolated in Inner Mongolia of China.BMC Vet Res. 2024 May 18;20(1):209. doi: 10.1186/s12917-024-04046-3. BMC Vet Res. 2024. PMID: 38760785 Free PMC article.
-
An Unwanted but Long-Known Company: Post-Viral Symptoms in the Context of Past Pandemics in Switzerland (and Beyond).Public Health Rev. 2024 Apr 8;45:1606966. doi: 10.3389/phrs.2024.1606966. eCollection 2024. Public Health Rev. 2024. PMID: 38651133 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

