Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair
- PMID: 15650050
- PMCID: PMC545844
- DOI: 10.1073/pnas.0407796102
Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair
Abstract
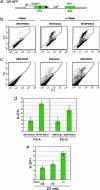
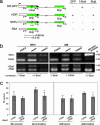
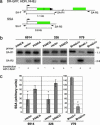
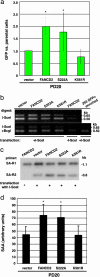
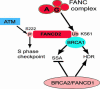
Fanconi anemia (FA) is a recessive disorder characterized by congenital abnormalities, progressive bone-marrow failure, and cancer susceptibility. Cells from FA patients are hypersensitive to agents that produce DNA crosslinks and, after treatment with these agents, have pronounced chromosome breakage and other cytogenetic abnormalities. Eight FANC genes have been cloned, and the encoded proteins interact in a common cellular pathway. DNA-damaging agents activate the monoubiquitination of FANCD2, resulting in its targeting to nuclear foci that also contain BRCA1 and BRCA2/FANCD1, proteins involved in homology-directed DNA repair. Given the interaction of the FANC proteins with BRCA1 and BRCA2, we tested whether cells from FA patients (groups A, G, and D2) and mouse Fanca-/- cells with a targeted mutation are impaired for this repair pathway. We find that both the upstream (FANCA and FANCG) and downstream (FANCD2) FA pathway components promote homology-directed repair of chromosomal double-strand breaks (DSBs). The FANCD2 monoubiquitination site is critical for normal levels of repair, whereas the ATM phosphorylation site is not. The defect in these cells, however, is mild, differentiating them from BRCA1 and BRCA2 mutant cells. Surprisingly, we provide evidence that these proteins, like BRCA1 but unlike BRCA2, promote a second DSB repair pathway involving homology, i.e., single-strand annealing. These results suggest an early role for the FANC proteins in homologous DSB repair pathway choice.
Figures
Similar articles
-
FANCD2 monoubiquitination and activation by hexavalent chromium [Cr(VI)] exposure: activation is not required for repair of Cr(VI)-induced DSBs.Mutat Res. 2006 Nov 7;610(1-2):21-30. doi: 10.1016/j.mrgentox.2006.06.009. Epub 2006 Aug 8. Mutat Res. 2006. PMID: 16893675 Free PMC article.
-
Function of the Fanconi anemia pathway in Fanconi anemia complementation group F and D1 cells.Exp Hematol. 2001 Dec;29(12):1448-55. doi: 10.1016/s0301-472x(01)00754-8. Exp Hematol. 2001. PMID: 11750104
-
The Chinese hamster FANCG/XRCC9 mutant NM3 fails to express the monoubiquitinated form of the FANCD2 protein, is hypersensitive to a range of DNA damaging agents and exhibits a normal level of spontaneous sister chromatid exchange.Carcinogenesis. 2001 Dec;22(12):1939-46. doi: 10.1093/carcin/22.12.1939. Carcinogenesis. 2001. PMID: 11751423
-
[Fanconi anemia: genes and function(s) revisited].Med Sci (Paris). 2005 Aug-Sep;21(8-9):730-6. doi: 10.1051/medsci/2005218-9730. Med Sci (Paris). 2005. PMID: 16115458 Review. French.
-
Molecular pathogenesis of fanconi anemia.Int J Hematol. 2002 Feb;75(2):123-8. doi: 10.1007/BF02982016. Int J Hematol. 2002. PMID: 11939257 Review.
Cited by
-
Fanconi anemia and dyskeratosis congenita/telomere biology disorders: Two inherited bone marrow failure syndromes with genomic instability.Front Oncol. 2022 Aug 25;12:949435. doi: 10.3389/fonc.2022.949435. eCollection 2022. Front Oncol. 2022. PMID: 36091172 Free PMC article. Review.
-
Adenine base editing efficiently restores the function of Fanconi anemia hematopoietic stem and progenitor cells.Nat Commun. 2022 Nov 12;13(1):6900. doi: 10.1038/s41467-022-34479-z. Nat Commun. 2022. PMID: 36371486 Free PMC article.
-
NuRD chromatin remodeling is required to repair exogenous DSBs in the Caenorhabditis elegans germline.bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613027. doi: 10.1101/2024.09.14.613027. bioRxiv. 2024. PMID: 39314477 Free PMC article. Preprint.
-
Causes and consequences of DNA double-stranded breaks in cardiovascular disease.Mol Cell Biochem. 2024 Oct 15. doi: 10.1007/s11010-024-05131-9. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39404936 Review.
-
Attenuating homologous recombination stimulates an AID-induced antileukemic effect.J Exp Med. 2013 May 6;210(5):1021-33. doi: 10.1084/jem.20121258. J Exp Med. 2013. PMID: 23589568 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

