Global expression changes resulting from loss of telomeric DNA in fission yeast
- PMID: 15642092
- PMCID: PMC549061
- DOI: 10.1186/gb-2004-6-1-r1
Global expression changes resulting from loss of telomeric DNA in fission yeast
Abstract
Background: Schizosaccharomyces pombe cells lacking the catalytic subunit of telomerase (encoded by trt1+) lose telomeric DNA and enter crisis, but rare survivors arise with either circular or linear chromosomes. Survivors with linear chromosomes have normal growth rates and morphology, but those with circular chromosomes have growth defects and are enlarged. We report the global gene-expression response of S. pombe to loss of trt1+.
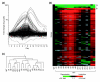
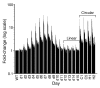
Results: Survivors with linear chromosomes had expression profiles similar to cells with native telomeres, whereas survivors with circular chromosomes showed continued upregulation of core environmental stress response (CESR) genes. In addition, survivors with circular chromosomes had altered expression of 51 genes compared to survivors with linear chromosomes, providing an expression signature. S. pombe progressing through crisis displayed two waves of altered gene expression. One coincided with crisis and consisted of around 110 genes, 44% of which overlapped with the CESR. The second was synchronized with the emergence of survivors and consisted of a single class of open reading frames (ORFs) with homology both to RecQ helicases and to dh repeats at centromeres targeted for heterochromatin formation via an RNA interference (RNAi) mechanism. Accumulation of transcript from the ORF was found not only in trt1- cells, but also in dcr1- and ago1- RNAi mutants, suggesting that RNAi may control its expression.
Conclusions: These results demonstrate a correlation between a state of cellular stress, short telomeres and growth defects in cells with circular chromosomes. A putative new RecQ helicase was expressed as survivors emerged and appears to be transcriptionally regulated by RNAi, suggesting that this mechanism operates at telomeres.
Figures


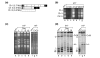

Similar articles
-
Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase.J Biol Chem. 2005 Feb 18;280(7):5249-57. doi: 10.1074/jbc.M412756200. Epub 2004 Dec 8. J Biol Chem. 2005. PMID: 15591066
-
Two modes of survival of fission yeast without telomerase.Science. 1998 Oct 16;282(5388):493-6. doi: 10.1126/science.282.5388.493. Science. 1998. PMID: 9774280
-
Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1.Nucleic Acids Res. 2006 Jan 10;34(1):78-88. doi: 10.1093/nar/gkj415. Print 2006. Nucleic Acids Res. 2006. PMID: 16407326 Free PMC article.
-
Protection and replication of telomeres in fission yeast.Biochem Cell Biol. 2009 Oct;87(5):747-58. doi: 10.1139/O09-037. Biochem Cell Biol. 2009. PMID: 19898524 Free PMC article. Review.
-
Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae.Mol Microbiol. 2006 Mar;59(5):1357-68. doi: 10.1111/j.1365-2958.2006.05026.x. Mol Microbiol. 2006. PMID: 16468981 Review.
Cited by
-
RNA interference and heterochromatin assembly.Cold Spring Harb Perspect Biol. 2011 Sep 1;3(9):a003731. doi: 10.1101/cshperspect.a003731. Cold Spring Harb Perspect Biol. 2011. PMID: 21441597 Free PMC article. Review.
-
RNA-induced initiation of transcriptional silencing (RITS) complex structure and function.RNA Biol. 2019 Sep;16(9):1133-1146. doi: 10.1080/15476286.2019.1621624. Epub 2019 Jun 18. RNA Biol. 2019. PMID: 31213126 Free PMC article. Review.
-
The telomere-linked helicase (TLH) gene family in Magnaporthe oryzae: revised gene structure reveals a novel TLH-specific protein motif.Curr Genet. 2009 Jun;55(3):253-62. doi: 10.1007/s00294-009-0240-3. Epub 2009 Apr 10. Curr Genet. 2009. PMID: 19360408
-
The 19S proteasome regulates subtelomere silencing and facultative heterochromatin formation in fission yeast.Curr Genet. 2018 Jun;64(3):741-752. doi: 10.1007/s00294-017-0792-6. Epub 2017 Dec 6. Curr Genet. 2018. PMID: 29214404
-
Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome.Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10542-7. doi: 10.1073/pnas.0703808104. Epub 2007 Jun 8. Proc Natl Acad Sci U S A. 2007. PMID: 17557836 Free PMC article.
References
-
- McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

