Brucella coopts the small GTPase Sar1 for intracellular replication
- PMID: 15632218
- PMCID: PMC547823
- DOI: 10.1073/pnas.0406873102
Brucella coopts the small GTPase Sar1 for intracellular replication
Abstract
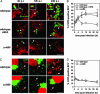
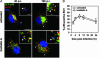
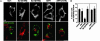
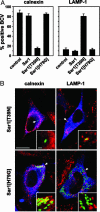
The pathogen Brucella abortus resides inside macrophages within a unique, replication-permissive organelle that is derived from the endoplasmic reticulum (ER). Although dependent on the Brucella type IV secretion system VirB, the mechanisms governing the biogenesis of this compartment remain elusive. Here, we investigated a putative role of the early secretory pathway in ER membrane accretion by the Brucella-containing vacuoles (BCVs). We show that BCVs interact with ER exit sites (ERES), and blockade of Sar1 activity, which disrupts ERES, prevents intracellular replication of Brucella. In cells expressing the dominant interfering form Sar1[T39N], BCVs do not acquire ER membranes, suggesting that they are unable to mature into replicative organelles. By contrast, treatments that block subsequent secretory events do not affect bacterial replication. We propose that Sar1-dependent ERES functions, but not subsequent secretory events, are essential for the biogenesis of the Brucella replicative compartment and, thus, bacterial replication. These results assign an essential role for Sar1 in pathogenesis of an intracellular bacterium.
Figures
Comment in
-
Bacterial subversion of the host secretory pathway.Proc Natl Acad Sci U S A. 2005 Feb 1;102(5):1271-2. doi: 10.1073/pnas.0409531101. Epub 2005 Jan 26. Proc Natl Acad Sci U S A. 2005. PMID: 15677328 Free PMC article. No abstract available.
Similar articles
-
Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum.J Exp Med. 2003 Aug 18;198(4):545-56. doi: 10.1084/jem.20030088. J Exp Med. 2003. PMID: 12925673 Free PMC article.
-
A T4SS Effector Targets Host Cell Alpha-Enolase Contributing to Brucella abortus Intracellular Lifestyle.Front Cell Infect Microbiol. 2016 Nov 16;6:153. doi: 10.3389/fcimb.2016.00153. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27900285 Free PMC article.
-
Postreplication Roles of the Brucella VirB Type IV Secretion System Uncovered via Conditional Expression of the VirB11 ATPase.mBio. 2016 Nov 29;7(6):e01730-16. doi: 10.1128/mBio.01730-16. mBio. 2016. PMID: 27899503 Free PMC article.
-
Organelle robbery: Brucella interactions with the endoplasmic reticulum.Curr Opin Microbiol. 2004 Feb;7(1):93-7. doi: 10.1016/j.mib.2003.11.001. Curr Opin Microbiol. 2004. PMID: 15036147 Review.
-
Surviving inside a macrophage: the many ways of Brucella.Res Microbiol. 2006 Mar;157(2):93-8. doi: 10.1016/j.resmic.2005.10.002. Epub 2005 Nov 9. Res Microbiol. 2006. PMID: 16364608 Review.
Cited by
-
Endoplasmic Reticulum Tubule Protein Reticulon 4 Associates with the Legionella pneumophila Vacuole and with Translocated Substrate Ceg9.Infect Immun. 2015 Sep;83(9):3479-89. doi: 10.1128/IAI.00507-15. Epub 2015 Jun 22. Infect Immun. 2015. PMID: 26099580 Free PMC article.
-
The enterohemorrhagic Escherichia coli effector protein NleF binds mammalian Tmp21.Vet Microbiol. 2013 May 31;164(1-2):164-70. doi: 10.1016/j.vetmic.2013.01.028. Epub 2013 Feb 4. Vet Microbiol. 2013. PMID: 23434013 Free PMC article.
-
Proteomic analysis of detergent resistant membrane domains during early interaction of macrophages with rough and smooth Brucella melitensis.PLoS One. 2014 Mar 18;9(3):e91706. doi: 10.1371/journal.pone.0091706. eCollection 2014. PLoS One. 2014. PMID: 24643124 Free PMC article.
-
The endoplasmic reticulum stress-mediated unfolded protein response protects against infection of goat endometrial epithelial cells by Trueperella pyogenes via autophagy.Virulence. 2022 Dec;13(1):122-136. doi: 10.1080/21505594.2021.2021630. Virulence. 2022. PMID: 34967271 Free PMC article.
-
The mechanism of chronic intracellular infection with Brucella spp.Front Cell Infect Microbiol. 2023 Apr 18;13:1129172. doi: 10.3389/fcimb.2023.1129172. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37143745 Free PMC article. Review.
References
-
- O'Callaghan, D., Cazevieille, C., Allardet-Servent, A., Boschiroli, M. L., Bourg, G., Foulongne, V., Frutos, P., Kulakov, Y. & Ramuz, M. (1999) Mol. Microbiol. 33, 1210–1220. - PubMed
-
- Comerci, D. J., Martinez-Lorenzo, M. J., Sieira, R., Gorvel, J. P. & Ugalde, R. A. (2001) Cell Microbiol. 3, 159–168. - PubMed
-
- Scales, S. J., Pepperkok, R. & Kreis, T. E. (1997) Cell 90, 1137–1148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

