Post-transcriptional gene silencing induced by short interfering RNAs in cultured transgenic plant cells
- PMID: 15629049
- PMCID: PMC5172445
- DOI: 10.1016/s1672-0229(04)02015-7
Post-transcriptional gene silencing induced by short interfering RNAs in cultured transgenic plant cells
Abstract
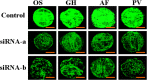
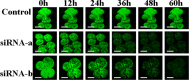
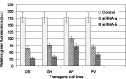
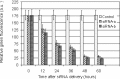
Short interfering RNA (siRNA) is widely used for studying post-transcriptional gene silencing and holds great promise as a tool for both identifying function of novel genes and validating drug targets. Two siRNA fragments (siRNA-a and -b), which were designed against different specific areas of coding region of the same target green fluorescent protein (GFP) gene, were used to silence GFP expression in cultured gfp transgenic cells of rice (Oryza sativa L.; OS), cotton (Gossypium hirsutum L.; GH), Fraser fir [Abies fraseri (Pursh) Poir; AF], and Virginia pine (Pinus virginiana Mill.; PV). Differential gene silencing was observed in the bombarded transgenic cells between two siRNAs, and these results were consistent with the inactivation of GFP confirmed by laser scanning microscopy, Northern blot, and siRNA analysis in tested transgenic cell cultures. These data suggest that siRNA-mediated gene inactivation can be the siRNA specific in different plant species. These results indicate that siRNA is a highly specific tool for targeted gene knockdown and for establishing siRNA-mediated gene silencing, which could be a reliable approach for large-scale screening of gene function and drug target validation.
Figures




Similar articles
-
Differential gene silencing induced by short interfering RNA in cultured pine cells associates with the cell cycle phase.Planta. 2006 Jun;224(1):53-60. doi: 10.1007/s00425-005-0190-z. Epub 2005 Dec 9. Planta. 2006. PMID: 16341704
-
Dexamethasone-inducible green fluorescent protein gene expression in transgenic plant cells.Genomics Proteomics Bioinformatics. 2004 Feb;2(1):15-23. doi: 10.1016/s1672-0229(04)02003-0. Genomics Proteomics Bioinformatics. 2004. PMID: 15629039 Free PMC article.
-
Inducible antisense-mediated post-transcriptional gene silencing in transgenic pine cells using green fluorescent protein as a visual marker.Plant Cell Physiol. 2005 Aug;46(8):1255-63. doi: 10.1093/pcp/pci134. Epub 2005 May 26. Plant Cell Physiol. 2005. PMID: 15919671
-
Post-transcriptional gene silencing in plants: a double-edged sword.Sci China Life Sci. 2016 Mar;59(3):271-6. doi: 10.1007/s11427-015-4972-7. Epub 2015 Dec 30. Sci China Life Sci. 2016. PMID: 26718356 Review.
-
Traffic into silence: endomembranes and post-transcriptional RNA silencing.EMBO J. 2014 May 2;33(9):968-80. doi: 10.1002/embj.201387262. Epub 2014 Mar 25. EMBO J. 2014. PMID: 24668229 Free PMC article. Review.
Cited by
-
Non-destructive analysis of the nuclei of transgenic living cells using laser tweezers and near-infrared raman spectroscopic technique.Genomics Proteomics Bioinformatics. 2005 Aug;3(3):169-78. doi: 10.1016/s1672-0229(05)03022-6. Genomics Proteomics Bioinformatics. 2005. PMID: 16487082 Free PMC article.
-
Engineering DNA nanostructures for siRNA delivery in plants.Nat Protoc. 2020 Sep;15(9):3064-3087. doi: 10.1038/s41596-020-0370-0. Epub 2020 Aug 17. Nat Protoc. 2020. PMID: 32807907 Free PMC article.
-
DNA nanostructures coordinate gene silencing in mature plants.Proc Natl Acad Sci U S A. 2019 Apr 9;116(15):7543-7548. doi: 10.1073/pnas.1818290116. Epub 2019 Mar 25. Proc Natl Acad Sci U S A. 2019. PMID: 30910954 Free PMC article.
-
RNA Interference for Functional Genomics and Improvement of Cotton (Gossypium sp.).Front Plant Sci. 2016 Feb 22;7:202. doi: 10.3389/fpls.2016.00202. eCollection 2016. Front Plant Sci. 2016. PMID: 26941765 Free PMC article. Review.
-
TDP-43 prevents retrotransposon activation in the Drosophila motor system through regulation of Dicer-2 activity.BMC Biol. 2020 Jul 3;18(1):82. doi: 10.1186/s12915-020-00816-1. BMC Biol. 2020. PMID: 32620127 Free PMC article.
References
-
- Schweizer P.J. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 2000;24:895–903. - PubMed
-
- Kanno T.S. Post-transcriptional gene silencing in cultured rice cells. Plant Cell Physiol. 2000;41:321–326. - PubMed
-
- Akashi H.M. Suppression of gene expression by RNA interference in cultured plant cells. Antisense Nucleic Acid Drug Dev. 2001;11:359–367. - PubMed
-
- Zamore P.D. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

