Side chain and backbone contributions of Phe508 to CFTR folding
- PMID: 15619636
- PMCID: PMC3516198
- DOI: 10.1038/nsmb881
Side chain and backbone contributions of Phe508 to CFTR folding
Abstract
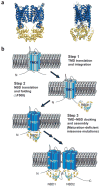
Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), an integral membrane protein, cause cystic fibrosis (CF). The most common CF-causing mutant, deletion of Phe508, fails to properly fold. To elucidate the role Phe508 plays in the folding of CFTR, missense mutations at this position were generated. Only one missense mutation had a pronounced effect on the stability and folding of the isolated domain in vitro. In contrast, many substitutions, including those of charged and bulky residues, disrupted folding of full-length CFTR in cells. Structures of two mutant nucleotide-binding domains (NBDs) reveal only local alterations of the surface near position 508. These results suggest that the peptide backbone plays a role in the proper folding of the domain, whereas the side chain plays a role in defining a surface of NBD1 that potentially interacts with other domains during the maturation of intact CFTR.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



Comment in
-
Arrest of CFTRDeltaF508 folding.Nat Struct Mol Biol. 2005 Jan;12(1):2-3. doi: 10.1038/nsmb0105-2. Nat Struct Mol Biol. 2005. PMID: 15689966 No abstract available.
Similar articles
-
The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR.Nat Struct Mol Biol. 2005 Jan;12(1):17-25. doi: 10.1038/nsmb882. Epub 2004 Dec 26. Nat Struct Mol Biol. 2005. PMID: 15619635
-
Deletion of Phe508 in the first nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator increases its affinity for the heat shock cognate 70 chaperone.FEBS J. 2009 Dec;276(23):7097-109. doi: 10.1111/j.1742-4658.2009.07421.x. Epub 2009 Oct 29. FEBS J. 2009. PMID: 19878303
-
Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure.J Biol Chem. 2005 Jan 14;280(2):1346-53. doi: 10.1074/jbc.M410968200. Epub 2004 Nov 3. J Biol Chem. 2005. PMID: 15528182
-
Misfolding of the cystic fibrosis transmembrane conductance regulator and disease.Biochemistry. 2008 Feb 12;47(6):1465-73. doi: 10.1021/bi702209s. Epub 2008 Jan 15. Biochemistry. 2008. PMID: 18193900 Review.
-
Dynamics intrinsic to cystic fibrosis transmembrane conductance regulator function and stability.Cold Spring Harb Perspect Med. 2013 Mar 1;3(3):a009522. doi: 10.1101/cshperspect.a009522. Cold Spring Harb Perspect Med. 2013. PMID: 23457292 Free PMC article. Review.
Cited by
-
Deletion of Phenylalanine 508 in the First Nucleotide-binding Domain of the Cystic Fibrosis Transmembrane Conductance Regulator Increases Conformational Exchange and Inhibits Dimerization.J Biol Chem. 2015 Sep 18;290(38):22862-78. doi: 10.1074/jbc.M115.641134. Epub 2015 Jul 6. J Biol Chem. 2015. PMID: 26149808 Free PMC article.
-
The role of cystic fibrosis transmembrane conductance regulator phenylalanine 508 side chain in ion channel gating.J Physiol. 2006 Apr 15;572(Pt 2):347-58. doi: 10.1113/jphysiol.2005.099457. Epub 2006 Feb 16. J Physiol. 2006. PMID: 16484308 Free PMC article.
-
Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal.Mol Biol Cell. 2007 Sep;18(9):3398-413. doi: 10.1091/mbc.e07-01-0046. Epub 2007 Jul 5. Mol Biol Cell. 2007. PMID: 17615300 Free PMC article.
-
Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis.Protein Sci. 2010 Oct;19(10):1932-47. doi: 10.1002/pro.480. Protein Sci. 2010. PMID: 20687163 Free PMC article.
-
The ABC protein turned chloride channel whose failure causes cystic fibrosis.Nature. 2006 Mar 23;440(7083):477-83. doi: 10.1038/nature04712. Nature. 2006. PMID: 16554808 Free PMC article. Review.
References
-
- Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–S45. - PubMed
-
- Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. - PubMed
-
- Tsui LC. The spectrum of cystic fibrosis mutations. Trends Genet. 1992;8:392–398. - PubMed
-
- Tsui LC. The cystic fibrosis transmembrane conductance regulator gene. Am J Respir Crit Care Med. 1995;151:S47–S53. - PubMed
-
- Cheng SH, et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

