Virtual screening of chemical libraries
- PMID: 15602552
- PMCID: PMC1360234
- DOI: 10.1038/nature03197
Virtual screening of chemical libraries
Abstract
Virtual screening uses computer-based methods to discover new ligands on the basis of biological structures. Although widely heralded in the 1970s and 1980s, the technique has since struggled to meet its initial promise, and drug discovery remains dominated by empirical screening. Recent successes in predicting new ligands and their receptor-bound structures, and better rates of ligand discovery compared to empirical screening, have re-ignited interest in virtual screening, which is now widely used in drug discovery, albeit on a more limited scale than empirical screening.
Conflict of interest statement
Figures
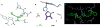

Similar articles
-
Comparative analysis of machine learning methods in ligand-based virtual screening of large compound libraries.Comb Chem High Throughput Screen. 2009 May;12(4):344-57. doi: 10.2174/138620709788167944. Comb Chem High Throughput Screen. 2009. PMID: 19442064 Review.
-
Synergies of virtual screening approaches.Mini Rev Med Chem. 2008 Aug;8(9):927-33. doi: 10.2174/138955708785132792. Mini Rev Med Chem. 2008. PMID: 18691150 Review.
-
How to benchmark methods for structure-based virtual screening of large compound libraries.Methods Mol Biol. 2012;819:187-95. doi: 10.1007/978-1-61779-465-0_13. Methods Mol Biol. 2012. PMID: 22183538
-
Structure-based virtual screening protocols.Curr Opin Drug Discov Devel. 2001 May;4(3):301-7. Curr Opin Drug Discov Devel. 2001. PMID: 11560062 Review.
-
The effect of ligand-based tautomer and protomer prediction on structure-based virtual screening.J Chem Inf Model. 2009 Dec;49(12):2742-8. doi: 10.1021/ci900364w. J Chem Inf Model. 2009. PMID: 19928753
Cited by
-
A Hybrid Docking and Machine Learning Approach to Enhance the Performance of Virtual Screening Carried out on Protein-Protein Interfaces.Int J Mol Sci. 2022 Nov 18;23(22):14364. doi: 10.3390/ijms232214364. Int J Mol Sci. 2022. PMID: 36430841 Free PMC article.
-
Implicit ligand theory: rigorous binding free energies and thermodynamic expectations from molecular docking.J Chem Phys. 2012 Sep 14;137(10):104106. doi: 10.1063/1.4751284. J Chem Phys. 2012. PMID: 22979849 Free PMC article.
-
Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.bioRxiv [Preprint]. 2024 Nov 17:2024.02.16.580675. doi: 10.1101/2024.02.16.580675. bioRxiv. 2024. PMID: 38405965 Free PMC article. Preprint.
-
Yuel: Improving the Generalizability of Structure-Free Compound-Protein Interaction Prediction.J Chem Inf Model. 2022 Feb 14;62(3):463-471. doi: 10.1021/acs.jcim.1c01531. Epub 2022 Feb 1. J Chem Inf Model. 2022. PMID: 35103472 Free PMC article.
-
Characterization of Entamoeba histolytica adenosine 5'-phosphosulfate (APS) kinase; validation as a target and provision of leads for the development of new drugs against amoebiasis.PLoS Negl Trop Dis. 2019 Aug 19;13(8):e0007633. doi: 10.1371/journal.pntd.0007633. eCollection 2019 Aug. PLoS Negl Trop Dis. 2019. PMID: 31425516 Free PMC article.
References
-
- Cohen SS. A strategy for the chemotherapy of infectious disease. Science. 1977;197:431–432. - PubMed
-
- Itzstein MV, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;36:418–423. - PubMed
-
- Varney MD, et al. Crystal-structure-based design and synthesis of Benz[cd]indole-containing inhibitors of thymidylate synthase. J. Med. Chem. 1992;35:663–676. - PubMed
-
- Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

