5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs
- PMID: 15572680
- PMCID: PMC533989
- DOI: 10.1128/MCB.24.24.10766-10776.2004
5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs
Abstract
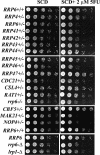
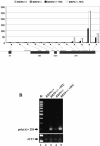
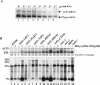
The antimetabolite 5-fluorouracil (5FU) is a widely used chemotherapeutic for the treatment of solid tumors. Although 5FU slows DNA synthesis by inhibiting the ability of thymidylate synthetase to produce dTMP, the drug also has significant effects on RNA metabolism. Recent genome-wide assays for 5FU-induced haploinsufficiency in Saccharomyces cerevisiae identified genes encoding components of the RNA processing exosome as potential targets of the drug. In this report, we used DNA microarrays to analyze the effect of 5FU on the yeast transcriptome and found that the drug causes the accumulation of polyadenylated fragments of the 27S rRNA precursor and that defects in the nuclear exoribonuclease Rrp6p enhance this effect. The size distribution of these RNAs and their sensitivity to Rrp6p suggest that they are normally degraded by the nuclear exosome and a 5'-3' exoribonuclease. Consistent with this hypothesis, 5FU inhibits the growth of RRP6 mutants with defects in the degradation function of the enzyme and it interferes with the degradation of an rRNA precursor. The detection of poly(A)(+) pre-RNAs in strains defective in various steps in ribosome biogenesis suggests that the production of poly(A)(+) pre-rRNAs may be a general result of defects in rRNA processing. These findings suggest that 5FU inhibits an exosome-dependent surveillance pathway that degrades polyadenylated precursor rRNAs.
Figures

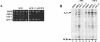
Similar articles
-
Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors.RNA. 2005 Oct;11(10):1571-8. doi: 10.1261/rna.2900205. Epub 2005 Aug 30. RNA. 2005. PMID: 16131592 Free PMC article.
-
Polyadenylation of rRNA in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8581-6. doi: 10.1073/pnas.0402888101. Epub 2004 Jun 1. Proc Natl Acad Sci U S A. 2004. PMID: 15173578 Free PMC article.
-
Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs.Mol Cell Biol. 2000 Jan;20(2):441-52. doi: 10.1128/MCB.20.2.441-452.2000. Mol Cell Biol. 2000. PMID: 10611222 Free PMC article.
-
Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination.Wiley Interdiscip Rev RNA. 2016 Jan-Feb;7(1):91-104. doi: 10.1002/wrna.1317. Epub 2015 Nov 26. Wiley Interdiscip Rev RNA. 2016. PMID: 26612606 Free PMC article. Review.
-
The exosome: a versatile RNA processing machine.Curr Biol. 1998 Mar 26;8(7):R238-40. doi: 10.1016/s0960-9822(98)70149-6. Curr Biol. 1998. PMID: 9583939 Review.
Cited by
-
Rapid identification of chemical genetic interactions in Saccharomyces cerevisiae.J Vis Exp. 2015 Apr 5;(98):e52345. doi: 10.3791/52345. J Vis Exp. 2015. PMID: 25867090 Free PMC article.
-
Retention of OsNMD3 in the cytoplasm disturbs protein synthesis efficiency and affects plant development in rice.J Exp Bot. 2014 Jul;65(12):3055-69. doi: 10.1093/jxb/eru150. Epub 2014 Apr 10. J Exp Bot. 2014. PMID: 24723395 Free PMC article.
-
Inhibiting eukaryotic ribosome biogenesis.BMC Biol. 2019 Jun 10;17(1):46. doi: 10.1186/s12915-019-0664-2. BMC Biol. 2019. PMID: 31182083 Free PMC article.
-
Progressive transcriptomic shifts in evolved yeast strains following gene knockout.iScience. 2024 Oct 21;27(11):111219. doi: 10.1016/j.isci.2024.111219. eCollection 2024 Nov 15. iScience. 2024. PMID: 39559754 Free PMC article.
-
Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae.EMBO J. 2006 Apr 5;25(7):1534-46. doi: 10.1038/sj.emboj.7601035. Epub 2006 Mar 16. EMBO J. 2006. PMID: 16541108 Free PMC article.
References
-
- Briggs, M. W., K. T. Burkard, and J. S. Butler. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273:13255-13263. - PubMed
-
- Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
