RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases
- PMID: 15545645
- PMCID: PMC1449118
- DOI: 10.1534/genetics.104.035964
RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases
Abstract
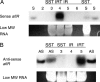
The versatility of RNA-dependent RNA polymerases (RDRPs) in eukaryotic gene silencing is perhaps best illustrated in the kingdom Fungi. Biochemical and genetic studies of Schizosaccharomyces pombe and Neurospora crassa show that these types of enzymes are involved in a number of fundamental gene-silencing processes, including heterochromatin regulation and RNA silencing in S. pombe and meiotic silencing and RNA silencing in N. crassa. Here we show that Aspergillus nidulans, another model fungus, does not require an RDRP for inverted repeat transgene (IRT)-induced RNA silencing. However, RDRP requirements may vary within the Aspergillus genus as genomic analysis indicates that A. nidulans, but not A. fumigatus or A. oryzae, has lost a QDE-1 ortholog, an RDRP associated with RNA silencing in N. crassa. We also provide evidence suggesting that 5' --> 3' transitive RNA silencing is not a significant aspect of A. nidulans IRT-RNA silencing. These results indicate a lack of conserved kingdom-wide requirements for RDRPs in fungal RNA silencing.
Figures




Similar articles
-
RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans.Eukaryot Cell. 2008 Feb;7(2):339-49. doi: 10.1128/EC.00355-07. Epub 2007 Dec 7. Eukaryot Cell. 2008. PMID: 18065653 Free PMC article.
-
The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa.Nucleic Acids Res. 2004 Apr 16;32(7):2123-8. doi: 10.1093/nar/gkh530. Print 2004. Nucleic Acids Res. 2004. PMID: 15090622 Free PMC article.
-
qiRNA is a new type of small interfering RNA induced by DNA damage.Nature. 2009 May 14;459(7244):274-7. doi: 10.1038/nature08041. Nature. 2009. PMID: 19444217 Free PMC article.
-
RNA interference in fungi: pathways, functions, and applications.Eukaryot Cell. 2011 Sep;10(9):1148-55. doi: 10.1128/EC.05109-11. Epub 2011 Jul 1. Eukaryot Cell. 2011. PMID: 21724934 Free PMC article. Review.
-
RNA interference pathways in fungi: mechanisms and functions.Annu Rev Microbiol. 2012;66:305-23. doi: 10.1146/annurev-micro-092611-150138. Epub 2012 Jun 28. Annu Rev Microbiol. 2012. PMID: 22746336 Free PMC article. Review.
Cited by
-
Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs.Eukaryot Cell. 2006 Apr;5(4):732-44. doi: 10.1128/EC.5.4.732-744.2006. Eukaryot Cell. 2006. PMID: 16607020 Free PMC article.
-
On the origin and functions of RNA-mediated silencing: from protists to man.Curr Genet. 2006 Aug;50(2):81-99. doi: 10.1007/s00294-006-0078-x. Epub 2006 May 12. Curr Genet. 2006. PMID: 16691418 Free PMC article. Review.
-
Mycovirus-encoded suppressors of RNA silencing: Possible allies or enemies in the use of RNAi to control fungal disease in crops.Front Fungal Biol. 2022 Oct 10;3:965781. doi: 10.3389/ffunb.2022.965781. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746227 Free PMC article. Review.
-
Inducible RNA Interference of brlAbeta in Aspergillus nidulans.Eukaryot Cell. 2008 Nov;7(11):2004-7. doi: 10.1128/EC.00142-08. Epub 2008 Aug 29. Eukaryot Cell. 2008. PMID: 18757565 Free PMC article.
-
The RNAi Machinery in the Fungus Fusarium fujikuroi Is Not Very Active in Synthetic Medium and Is Related to Transposable Elements.Noncoding RNA. 2024 May 16;10(3):31. doi: 10.3390/ncrna10030031. Noncoding RNA. 2024. PMID: 38804363 Free PMC article.
References
-
- Beclin, C., S. Boutet, P. Waterhouse and H. Vaucheret, 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688. - PubMed
-
- Bernstein, E., A. A. Caudy, S. M. Hammond and G. J. Hannon, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

