siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis
- PMID: 15545622
- PMCID: PMC534648
- DOI: 10.1101/gad.1217304
siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis
Abstract
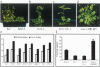
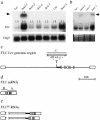
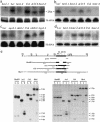
Allelic variation in FLOWERING LOCUS C (FLC), a central repressor of flowering, contributes to natural differences in flowering behavior among Arabidopsis accessions. The weak nature of the FLC allele in the Ler accession is due to low levels of FLC RNA resulting, through an unknown mechanism, from a transposable element inserted in an intron of FLC. Here we show that the transposable element renders FLC-Ler subject to repressive chromatin modifications mediated by short interfering RNAs generated from homologous transposable elements in the genome. Our studies have general implications for the role of transposable elements in eukaryotic gene expression and evolution.
Figures

Similar articles
-
Small RNA-directed epigenetic natural variation in Arabidopsis thaliana.PLoS Genet. 2008 Apr 25;4(4):e1000056. doi: 10.1371/journal.pgen.1000056. PLoS Genet. 2008. PMID: 18437202 Free PMC article.
-
Analysis of the molecular basis of flowering time variation in Arabidopsis accessions.Plant Physiol. 2003 Jun;132(2):1107-14. doi: 10.1104/pp.103.021212. Epub 2003 May 22. Plant Physiol. 2003. PMID: 12805638 Free PMC article.
-
Small RNA-mediated chromatin silencing directed to the 3' region of the Arabidopsis gene encoding the developmental regulator, FLC.Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3633-8. doi: 10.1073/pnas.0611459104. Epub 2007 Feb 20. Proc Natl Acad Sci U S A. 2007. PMID: 17360694 Free PMC article.
-
Chromatin regulation of flowering.Trends Plant Sci. 2012 Sep;17(9):556-62. doi: 10.1016/j.tplants.2012.05.001. Epub 2012 Jun 2. Trends Plant Sci. 2012. PMID: 22658650 Review.
-
siRNAs and DNA methylation: seedy epigenetics.Trends Plant Sci. 2010 Apr;15(4):204-10. doi: 10.1016/j.tplants.2010.01.002. Epub 2010 Feb 1. Trends Plant Sci. 2010. PMID: 20129810 Review.
Cited by
-
The contribution of transposable elements to transcriptional novelty in plants: the FLC affair.Transcription. 2020 Jun-Aug;11(3-4):192-198. doi: 10.1080/21541264.2020.1803031. Epub 2020 Aug 12. Transcription. 2020. PMID: 32783496 Free PMC article. Review.
-
A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing.Plant Cell. 2007 Sep;19(9):2929-39. doi: 10.1105/tpc.107.051821. Epub 2007 Sep 21. Plant Cell. 2007. PMID: 17890374 Free PMC article.
-
Genome-wide association study revealed that the TaGW8 gene was associated with kernel size in Chinese bread wheat.Sci Rep. 2019 Feb 25;9(1):2702. doi: 10.1038/s41598-019-38570-2. Sci Rep. 2019. PMID: 30804359 Free PMC article.
-
Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs.Genome Res. 2009 Jan;19(1):42-56. doi: 10.1101/gr.078196.108. Epub 2008 Nov 26. Genome Res. 2009. PMID: 19037014 Free PMC article.
-
Small RNA-directed epigenetic natural variation in Arabidopsis thaliana.PLoS Genet. 2008 Apr 25;4(4):e1000056. doi: 10.1371/journal.pgen.1000056. PLoS Genet. 2008. PMID: 18437202 Free PMC article.
References
-
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281-297. - PubMed
-
- Boutet S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. 2003. Arabidopsis HEN1. A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843-848. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
