CREB binding protein functions during successive stages of eye development in Drosophila
- PMID: 15514061
- PMCID: PMC1448854
- DOI: 10.1534/genetics.104.029850
CREB binding protein functions during successive stages of eye development in Drosophila
Abstract
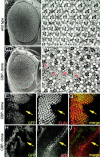
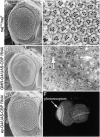
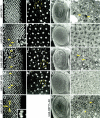
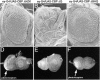
During the development of the compound eye of Drosophila several signaling pathways exert both positive and inhibitory influences upon an array of nuclear transcription factors to produce a near-perfect lattice of unit eyes or ommatidia. Individual cells within the eye are exposed to many extracellular signals, express multiple surface receptors, and make use of a large complement of cell-subtype-specific DNA-binding transcription factors. Despite this enormous complexity, each cell will make the correct developmental choice and adopt the appropriate cell fate. How this process is managed remains a poorly understood paradigm. Members of the CREB binding protein (CBP)/p300 family have been shown to influence development by (1) acting as bridging molecules between the basal transcriptional machinery and specific DNA-binding transcription factors, (2) physically interacting with terminal members of signaling cascades, (3) acting as transcriptional coactivators of downstream target genes, and (4) playing a key role in chromatin remodeling. In a screen for new genes involved in eye development we have identified the Drosophila homolog of CBP as a key player in both eye specification and cell fate determination. We have used a variety of approaches to define the role of CBP in eye development on a cell-by-cell basis.
Figures
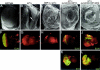

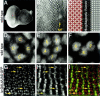
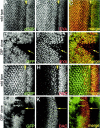
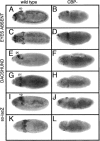


Similar articles
-
A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye.Genetics. 2005 Dec;171(4):1655-72. doi: 10.1534/genetics.105.045450. Epub 2005 Jul 5. Genetics. 2005. PMID: 15998717 Free PMC article.
-
Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers.Mol Cell Biol. 2000 Dec;20(24):9317-30. doi: 10.1128/MCB.20.24.9317-9330.2000. Mol Cell Biol. 2000. PMID: 11094082 Free PMC article.
-
A function of CBP as a transcriptional co-activator during Dpp signalling.EMBO J. 1999 Mar 15;18(6):1630-41. doi: 10.1093/emboj/18.6.1630. EMBO J. 1999. PMID: 10075933 Free PMC article.
-
Transcriptional coregulators in development.Science. 1999 Apr 23;284(5414):606-9. doi: 10.1126/science.284.5414.606. Science. 1999. PMID: 10213677 Review.
-
United colours of chromatin? Developmental genome organisation in flies.Biochem Soc Trans. 2019 Apr 30;47(2):691-700. doi: 10.1042/BST20180605. Epub 2019 Mar 22. Biochem Soc Trans. 2019. PMID: 30902925 Review.
Cited by
-
Cis-regulatory signatures of orthologous stress-associated bZIP transcription factors from rice, sorghum and Arabidopsis based on phylogenetic footprints.BMC Genomics. 2012 Sep 20;13:497. doi: 10.1186/1471-2164-13-497. BMC Genomics. 2012. PMID: 22992304 Free PMC article.
-
A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye.Genetics. 2005 Dec;171(4):1655-72. doi: 10.1534/genetics.105.045450. Epub 2005 Jul 5. Genetics. 2005. PMID: 15998717 Free PMC article.
-
A screen for X-linked mutations affecting Drosophila photoreceptor differentiation identifies Casein kinase 1α as an essential negative regulator of wingless signaling.Genetics. 2012 Feb;190(2):601-16. doi: 10.1534/genetics.111.133827. Epub 2011 Nov 17. Genetics. 2012. PMID: 22095083 Free PMC article.
-
Glaucoma-TrEl: A web-based interactive database to build evidence-based hypotheses on the role of trace elements in glaucoma.BMC Res Notes. 2022 Nov 18;15(1):348. doi: 10.1186/s13104-022-06210-0. BMC Res Notes. 2022. PMID: 36401306 Free PMC article.
-
Drosophila CtBP regulates proliferation and differentiation of eye precursors and complexes with Eyeless, Dachshund, Dan, and Danr during eye and antennal development.Dev Dyn. 2010 Sep;239(9):2367-85. doi: 10.1002/dvdy.22380. Dev Dyn. 2010. PMID: 20730908 Free PMC article.
References
-
- Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet et al., 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262: 157–162. - PubMed
-
- Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka et al., 1997. a Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386: 735–738. - PubMed
-
- Akimaru, H., D. X. Hou and S. Ishii, 1997. b Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet. 17: 211–214. - PubMed
-
- Arany, Z., W. R. Sellers, D. M. Livingston and R. Eckner, 1994. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77: 799–800. - PubMed
-
- Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine et al., 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89: 1175–1184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

