Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting
- PMID: 15511219
- PMCID: PMC1134928
- DOI: 10.1042/BJ20041227
Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting
Abstract
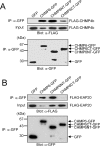
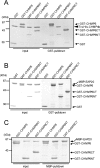
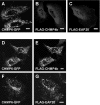
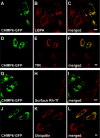
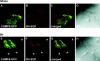
CHMP6 (charged multivesicular body protein 6) is a human orthologue of yeast Vps (vacuolar protein sorting) 20, a component of ESCRT (endosomal sorting complex required for transport)-III. Various CHMP6 orthologues in organisms ranging from yeast to humans contain the N-myristoylation consensus sequence at each N-terminus. Metabolic labelling of HEK-293 (human embryonic kidney) cells showed the incorporation of [3H]myristate into CHMP6 fused C-terminally to GFP (green fluorescent protein) (CHMP6-GFP). Interactions of CHMP6 with another ESCRT-III component CHMP4b/Shax [Snf7 (sucrose non-fermenting 7) homologue associated with Alix] 1, one of three paralogues of human Vps32/Snf7, and with EAP20 (ELL-associated protein 20), a human counterpart of yeast Vps25 and component of ESCRT-II, were observed by co-immunoprecipitation of epitope-tagged proteins expressed in HEK-293 cells. The in vitro pull-down assays using their recombinant proteins purified from Escherichia coli demonstrated direct physical interactions which were mediated by the N-terminal basic half of CHMP6. Overexpressed CHMP6-GFP in HeLa cells exhibited a punctate distribution throughout the cytoplasm especially in the perinuclear area, as revealed by fluorescence microscopic analysis. Accumulation of LBPA (lysobisphosphatidic acid), a major phospholipid in internal vesicles of an MVB (multivesicular body), was observed in the CHMP6-GFP-localizing area. FLAG-tagged EAP20 distributed diffusely, but exhibited a punctate distribution on co-expression with CHMP6-GFP. Overexpression of CHMP6-GFP caused reduction of transferrin receptors on the plasma membrane surface, but caused their accumulation in the cytoplasm. Ubiquitinated proteins and endocytosed EGF continuously accumulated in CHMP6-GFP-expressing cells. These results suggest that CHMP6 acts as an acceptor for ESCRT-II on endosomal membranes and regulates cargo sorting.
Figures

Similar articles
-
CHMP7, a novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway.Biochem J. 2006 Nov 15;400(1):23-32. doi: 10.1042/BJ20060897. Biochem J. 2006. PMID: 16856878 Free PMC article.
-
Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation.Dev Cell. 2008 Oct;15(4):578-89. doi: 10.1016/j.devcel.2008.08.013. Dev Cell. 2008. PMID: 18854142
-
The penta-EF-hand protein ALG-2 interacts directly with the ESCRT-I component TSG101, and Ca2+-dependently co-localizes to aberrant endosomes with dominant-negative AAA ATPase SKD1/Vps4B.Biochem J. 2005 Nov 1;391(Pt 3):677-85. doi: 10.1042/BJ20050398. Biochem J. 2005. PMID: 16004603 Free PMC article.
-
ESCRT proteins in physiology and disease.Exp Cell Res. 2009 May 15;315(9):1619-26. doi: 10.1016/j.yexcr.2008.10.013. Epub 2008 Oct 28. Exp Cell Res. 2009. PMID: 19013455 Review.
-
Endosomal and non-endosomal functions of ESCRT proteins.Trends Cell Biol. 2006 Jun;16(6):317-26. doi: 10.1016/j.tcb.2006.04.004. Epub 2006 May 22. Trends Cell Biol. 2006. PMID: 16716591 Review.
Cited by
-
Shaping development with ESCRTs.Nat Cell Biol. 2011 Dec 22;14(1):38-45. doi: 10.1038/ncb2381. Nat Cell Biol. 2011. PMID: 22193162 Review.
-
Septin Remodeling During Mammalian Cytokinesis.Front Cell Dev Biol. 2021 Nov 4;9:768309. doi: 10.3389/fcell.2021.768309. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34805175 Free PMC article. Review.
-
No strings attached: the ESCRT machinery in viral budding and cytokinesis.J Cell Sci. 2009 Jul 1;122(Pt 13):2167-77. doi: 10.1242/jcs.028308. J Cell Sci. 2009. PMID: 19535732 Free PMC article.
-
Membrane budding.Cell. 2010 Dec 10;143(6):875-87. doi: 10.1016/j.cell.2010.11.030. Cell. 2010. PMID: 21145455 Free PMC article. Review.
-
ESCRT Requirements for Murine Leukemia Virus Release.Viruses. 2016 Apr 18;8(4):103. doi: 10.3390/v8040103. Viruses. 2016. PMID: 27096867 Free PMC article.
References
-
- Maxfield F. R., McGraw T. E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 2004;5:121–132. - PubMed
-
- Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature (London) 1998;392:193–197. - PubMed
-
- Mukherjee S., Maxfield F. R. Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic. 2000;1:203–211. - PubMed
-
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. - PubMed
-
- Di Fiore P. P., Polo S., Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell Biol. 2003;4:491–497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

