Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40
- PMID: 15500443
- PMCID: PMC1134863
- DOI: 10.1042/BJ20041050
Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40
Abstract
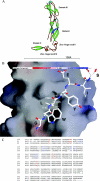
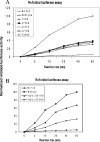
Ydj1 is the major type I Hsp40 (heat-shock protein 40) family member in yeast. Ydj1 can pair with yeast Hsp70 Ssa1 to facilitate protein translocation and protein folding. Ydj1 itself can also function as a molecular chaperone to bind the non-native polypeptides and suppress protein aggregations in vitro. The crystal structure of Ydj1 complexed with its peptide substrate GWLYEIS reveals that a hydrophobic pocket located on Ydj1 domain I may play a major role in mediating the interactions between Ydj1 and the peptide substrate. To understand the mechanism by which Ydj1 interacts with non-native polypeptide, we have mutated the residues forming the hydrophobic pocket, based on the structural information. We have also constructed deletion mutations of the zinc-finger motifs within Ydj1. We have examined the functional consequences of these Ydj1 mutants by in vivo and in vitro assays. The results indicated that the hydrophobic pocket located on Ydj1 plays a critical role in its molecular chaperone activity by mediating interactions with the non-native polypeptides.
Figures



Similar articles
-
The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding.J Biol Chem. 1998 Mar 6;273(10):5970-8. doi: 10.1074/jbc.273.10.5970. J Biol Chem. 1998. PMID: 9488737
-
The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.Structure. 2003 Dec;11(12):1475-83. doi: 10.1016/j.str.2003.10.012. Structure. 2003. PMID: 14656432
-
Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1.Biochem J. 2002 Jan 1;361(Pt 1):27-34. doi: 10.1042/0264-6021:3610027. Biochem J. 2002. PMID: 11743879 Free PMC article.
-
Role of J-domain Proteins in Yeast Physiology and Protein Quality Control.J Mol Biol. 2024 Jul 15;436(14):168484. doi: 10.1016/j.jmb.2024.168484. Epub 2024 Feb 7. J Mol Biol. 2024. PMID: 38331212 Review.
-
Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation.Crit Rev Biochem Mol Biol. 2007 Mar-Apr;42(2):95-111. doi: 10.1080/10409230701322298. Crit Rev Biochem Mol Biol. 2007. PMID: 17453917 Review.
Cited by
-
Mechanisms of the Hsp70 chaperone system.Biochem Cell Biol. 2010 Apr;88(2):291-300. doi: 10.1139/o09-175. Biochem Cell Biol. 2010. PMID: 20453930 Free PMC article. Review.
-
Mammalian Fe-S proteins: definition of a consensus motif recognized by the co-chaperone HSC20.Metallomics. 2016 Oct 1;8(10):1032-1046. doi: 10.1039/c6mt00167j. Metallomics. 2016. PMID: 27714045 Free PMC article. Review.
-
Curing of yeast [URE3] prion by the Hsp40 cochaperone Ydj1p is mediated by Hsp70.Genetics. 2009 Jan;181(1):129-37. doi: 10.1534/genetics.108.098699. Epub 2008 Nov 17. Genetics. 2009. PMID: 19015537 Free PMC article.
-
Role of the J Domain Protein Family in the Survival and Pathogenesis of Plasmodium falciparum.Adv Exp Med Biol. 2021;1340:97-123. doi: 10.1007/978-3-030-78397-6_4. Adv Exp Med Biol. 2021. PMID: 34569022 Review.
-
Structural insights into the chaperone activity of the 40-kDa heat shock protein DnaJ: binding and remodeling of a native substrate.J Biol Chem. 2013 May 24;288(21):15065-74. doi: 10.1074/jbc.M112.430595. Epub 2013 Apr 11. J Biol Chem. 2013. PMID: 23580641 Free PMC article.
References
-
- Bukau B., Horwich A. L. The Hsp70 and Hsp60 chaperone machines. Cell (Cambridge, Mass.) 1998;92:351–366. - PubMed
-
- Hartl F. U. Molecular chaperones in cellular protein folding. Nature (London) 1996;381:571–579. - PubMed
-
- Hartl F. U., Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. - PubMed
-
- Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature (London) 1992;356:683–689. - PubMed
-
- Schmid D., Baici A., Gehring H., Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

