HSF4 is required for normal cell growth and differentiation during mouse lens development
- PMID: 15483628
- PMCID: PMC524399
- DOI: 10.1038/sj.emboj.7600435
HSF4 is required for normal cell growth and differentiation during mouse lens development
Abstract
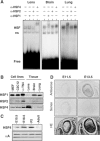
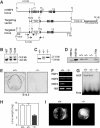
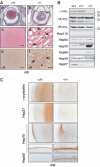
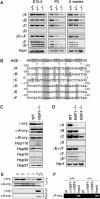
The heat shock transcription factor (HSF) family consists of three members in mammals and regulates expression of heat shock genes via a heat shock element. HSF1 and HSF2 are required for some developmental processes, but it is unclear how they regulate these processes. To elucidate the mechanisms of developmental regulation by HSFs, we generated mice in which the HSF4 gene is mutated. HSF4-null mice had cataract with abnormal lens fiber cells containing inclusion-like structures, probably due to decreased expression of gamma-crystallin, which maintains protein stability. Furthermore, we found increased proliferation and premature differentiation of the mutant lens epithelial cells, which is associated with increased expression of growth factors, FGF-1, FGF-4, and FGF-7. Unexpectedly, HSF1 competed with HSF4 for the expression of FGFs not only in the lens but also in other tissues. These findings reveal the lens-specific role of HSF4, which activates gamma-crystallin genes, and also indicate that HSF1 and HSF4 are involved in regulating expression of growth factor genes, which are essential for cell growth and differentiation.
Figures
Similar articles
-
Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation.Genesis. 2004 Dec;40(4):205-17. doi: 10.1002/gene.20087. Genesis. 2004. PMID: 15593327
-
Identification of vimentin as a novel target of HSF4 in lens development and cataract by proteomic analysis.Invest Ophthalmol Vis Sci. 2010 Jan;51(1):396-404. doi: 10.1167/iovs.09-3772. Epub 2009 Jul 23. Invest Ophthalmol Vis Sci. 2010. PMID: 19628735
-
Developmentally dictated expression of heat shock factors: exclusive expression of HSF4 in the postnatal lens and its specific interaction with alphaB-crystallin heat shock promoter.J Biol Chem. 2004 Oct 22;279(43):44497-503. doi: 10.1074/jbc.M405813200. Epub 2004 Aug 12. J Biol Chem. 2004. PMID: 15308659
-
Transcriptional regulation of small HSP-HSF1 and beyond.Int J Biochem Cell Biol. 2012 Oct;44(10):1593-612. doi: 10.1016/j.biocel.2012.06.012. Epub 2012 Jun 29. Int J Biochem Cell Biol. 2012. PMID: 22750029 Review.
-
Heat shock factors at a crossroad between stress and development.Ann N Y Acad Sci. 2007 Oct;1113:15-27. doi: 10.1196/annals.1391.005. Epub 2007 May 4. Ann N Y Acad Sci. 2007. PMID: 17483205 Review.
Cited by
-
A novel missense mutation in the HSF4 gene of giant pandas with senile congenital cataracts.Sci Rep. 2021 Mar 8;11(1):5411. doi: 10.1038/s41598-021-84741-5. Sci Rep. 2021. PMID: 33686159 Free PMC article.
-
Genetic and epigenetic mechanisms of gene regulation during lens development.Prog Retin Eye Res. 2007 Nov;26(6):555-97. doi: 10.1016/j.preteyeres.2007.07.002. Epub 2007 Jul 28. Prog Retin Eye Res. 2007. PMID: 17905638 Free PMC article. Review.
-
Heat shock factors: integrators of cell stress, development and lifespan.Nat Rev Mol Cell Biol. 2010 Aug;11(8):545-55. doi: 10.1038/nrm2938. Epub 2010 Jul 14. Nat Rev Mol Cell Biol. 2010. PMID: 20628411 Free PMC article. Review.
-
Molecular Mechanisms of Heat Shock Factors in Cancer.Cells. 2020 May 12;9(5):1202. doi: 10.3390/cells9051202. Cells. 2020. PMID: 32408596 Free PMC article. Review.
-
Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes.PLoS One. 2014 Jul 21;9(7):e102825. doi: 10.1371/journal.pone.0102825. eCollection 2014. PLoS One. 2014. PMID: 25047803 Free PMC article.
References
-
- Bhat SP (2003) Crystallins, genes and cataract. Prog Drug Res 60: 205–262 - PubMed
-
- Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X (2002) Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet 31: 276–278 - PubMed
-
- Burgess WH, Maciag T (1989) The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem 58: 575–606 - PubMed
-
- Christians E, Davis AA, Thomas SD, Benjamin IJ (2000) Maternal effect of Hsf1 on reproductive success. Nature 407: 693–694 - PubMed
-
- Fagerholm PP, Philipson BT, Lindstrom B (1981) Normal human lens—the distribution of protein. Exp Eye Res 33: 615–620 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

