Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization
- PMID: 15479947
- PMCID: PMC525687
- DOI: 10.1101/gr.2896904
Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization
Erratum in
- Genome Res. 2004 Dec;14(12):2510
Abstract
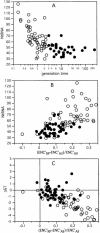
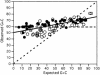
The selection-mutation-drift theory of codon usage plays a major role in the theory of molecular evolution by explaining the co-evolution of codon usage bias and tRNA content in the framework of translation optimization. Because most studies have focused only on codon usage, we analyzed the tRNA gene pool of 102 bacterial species. We show that as minimal generation times get shorter, the genomes contain more tRNA genes, but fewer anticodon species. Surprisingly, despite the wide G+C variation of bacterial genomes these anticodons are the same in most genomes. This suggests an optimization of the translation machinery to use a small subset of optimal codons and anticodons in fast-growing bacteria and in highly expressed genes. As a result, the overrepresented codons in highly expressed genes tend to be the same in very different genomes to match the same most-frequent anticodons. This is particularly important in fast-growing bacteria, which have higher codon usage bias in these genes. Three models were tested to understand the choice of codons recognized by the same anticodons, all providing significant fit, but under different classes of genes and genomes. Thus, co-evolution of tRNA gene composition and codon usage bias in genomes seen from tRNA's point of view agrees with the selection-mutation-drift theory. However, it suggests a much more universal trend in the evolution of anticodon and codon choice than previously thought. It also provides new evidence that a selective force for the optimization of the translation machinery is the maximization of growth.
Figures
Similar articles
-
Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage.Mol Biol Evol. 2008 Nov;25(11):2279-91. doi: 10.1093/molbev/msn173. Epub 2008 Aug 6. Mol Biol Evol. 2008. PMID: 18687657
-
Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes.Gene. 2005 Jan 17;345(1):13-20. doi: 10.1016/j.gene.2004.11.019. Epub 2004 Dec 19. Gene. 2005. PMID: 15716092
-
The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria.Mol Biol Evol. 2010 Sep;27(9):2129-40. doi: 10.1093/molbev/msq102. Epub 2010 Apr 19. Mol Biol Evol. 2010. PMID: 20403966
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
-
tRNA's wobble decoding of the genome: 40 years of modification.J Mol Biol. 2007 Feb 9;366(1):1-13. doi: 10.1016/j.jmb.2006.11.046. Epub 2006 Nov 15. J Mol Biol. 2007. PMID: 17187822 Review.
Cited by
-
Codon Usage is Influenced by Compositional Constraints in Genes Associated with Dementia.Front Genet. 2022 Aug 9;13:884348. doi: 10.3389/fgene.2022.884348. eCollection 2022. Front Genet. 2022. PMID: 36017501 Free PMC article.
-
Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency.PLoS One. 2012;7(3):e33547. doi: 10.1371/journal.pone.0033547. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432034 Free PMC article.
-
Depletion of Shine-Dalgarno Sequences Within Bacterial Coding Regions Is Expression Dependent.G3 (Bethesda). 2016 Nov 8;6(11):3467-3474. doi: 10.1534/g3.116.032227. G3 (Bethesda). 2016. PMID: 27605518 Free PMC article.
-
An assessment of the impacts of molecular oxygen on the evolution of proteomes.Mol Biol Evol. 2008 Sep;25(9):1931-42. doi: 10.1093/molbev/msn142. Epub 2008 Jun 25. Mol Biol Evol. 2008. PMID: 18579552 Free PMC article.
-
The cost of wobble translation in fungal mitochondrial genomes: integration of two traditional hypotheses.BMC Evol Biol. 2008 Jul 19;8:211. doi: 10.1186/1471-2148-8-211. BMC Evol Biol. 2008. PMID: 18638409 Free PMC article.
References
-
- Baumann, P., Baumann, L., Lai, C.-H., and Rouhbakhsh, D. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu. Rev. Microbiol. 49: 55-94. - PubMed
-
- Berg, O.G. and Kurland, C.G. 1997. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol. 270: 544-550. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
