Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series
- PMID: 15459747
- PMCID: PMC1299150
- DOI: 10.1038/sj.embor.7400250
Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series
Abstract
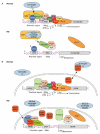
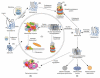
Huntington's disease (HD) is a late-onset neurodegenerative disorder that is caused by a CAG repeat expansion in the IT15 gene, which results in a long stretch of polyglutamine close to the amino-terminus of the HD protein huntingtin (htt). The normal function of htt, and the molecular mechanisms that contribute to the disease pathogenesis, are in the process of being elucidated. In this review, we outline the potential functions of htt as defined by the proteins with which it has been found to interact. We then focus on evidence that supports a role for transcriptional dysfunction and impaired protein folding and degradation as early events in disease pathogenesis.
Figures

Similar articles
-
Huntington's Disease.Cold Spring Harb Perspect Biol. 2011 Jun 1;3(6):a007476. doi: 10.1101/cshperspect.a007476. Cold Spring Harb Perspect Biol. 2011. PMID: 21441583 Free PMC article. Review.
-
Huntington's Disease: Mechanisms of Pathogenesis and Therapeutic Strategies.Cold Spring Harb Perspect Med. 2017 Jul 5;7(7):a024240. doi: 10.1101/cshperspect.a024240. Cold Spring Harb Perspect Med. 2017. PMID: 27940602 Free PMC article. Review.
-
PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD).Cell Cycle. 2015;14(11):1716-29. doi: 10.1080/15384101.2015.1033595. Cell Cycle. 2015. PMID: 25927346 Free PMC article.
-
Lessons from animal models of Huntington's disease.Trends Genet. 2002 Apr;18(4):202-9. doi: 10.1016/s0168-9525(01)02625-7. Trends Genet. 2002. PMID: 11932021 Review.
-
Huntington's disease: translating a CAG repeat into a pathogenic mechanism.Curr Opin Neurobiol. 1996 Oct;6(5):638-43. doi: 10.1016/s0959-4388(96)80097-3. Curr Opin Neurobiol. 1996. PMID: 8937828 Review.
Cited by
-
Caspase-6 does not contribute to the proteolysis of mutant huntingtin in the HdhQ150 knock-in mouse model of Huntington's disease.PLoS Curr. 2012 Jul 16;4:e4fd085bfc9973. doi: 10.1371/4fd085bfc9973. PLoS Curr. 2012. PMID: 22919566 Free PMC article. No abstract available.
-
Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease.J Biol Chem. 2010 Mar 19;285(12):8808-23. doi: 10.1074/jbc.M109.075028. Epub 2010 Jan 19. J Biol Chem. 2010. PMID: 20086007 Free PMC article.
-
Amphiphilic Nanocarrier Systems for Curcumin Delivery in Neurodegenerative Disorders.Medicines (Basel). 2018 Nov 23;5(4):126. doi: 10.3390/medicines5040126. Medicines (Basel). 2018. PMID: 30477087 Free PMC article. Review.
-
Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype.PLoS One. 2009 Sep 30;4(9):e7232. doi: 10.1371/journal.pone.0007232. PLoS One. 2009. PMID: 19789634 Free PMC article.
-
Mutant huntingtin inhibits the mitochondrial unfolded protein response by impairing ABCB10 mRNA stability.Biochim Biophys Acta Mol Basis Dis. 2019 Jun 1;1865(6):1428-1435. doi: 10.1016/j.bbadis.2019.02.015. Epub 2019 Feb 23. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 30802639 Free PMC article.
References
-
- Bates G (2003) Huntingtin aggregation and toxicity in Huntington's disease. Lancet 361: 1642–1644 - PubMed
-
- Bates GP, Murphy KP (2002) in Huntington's Disease (eds Bates GP, Harper PS, Jones AL) 387–426. Oxford, UK: Oxford University Press
-
- Bates GP, Harper PS, Jones AL (eds) (2002) Huntington's Disease. Oxford, UK: Oxford University Press.
-
- Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin–proteasome system by protein aggregation. Science 292: 1552–1555 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

