Stimulatory G protein directly regulates hypertrophic differentiation of growth plate cartilage in vivo
- PMID: 15459318
- PMCID: PMC522030
- DOI: 10.1073/pnas.0405091101
Stimulatory G protein directly regulates hypertrophic differentiation of growth plate cartilage in vivo
Abstract
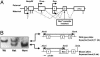
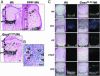
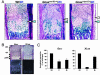
Stimulatory heterotrimeric G protein (Gs) transduces signals from various cell-surface receptors to adenylyl cyclases, which generate cAMP. The alpha subunit of Gs (Gsalpha) is encoded by GNAS (Gnas in mice), and heterozygous Gsalpha inactivating mutations lead to Albright hereditary osteodystrophy. The in vivo role of Gsalpha in skeletogenesis is largely unknown, because of early embryonic lethality of mice with disruption of Gnas exon 2 (Gnas(E2-/E2-)) and the absence of easily detectable phenotypes in growth plate chondrocytes of heterozygous mutant mice (Gnas(+/E2-)). We generated chimeric mice containing wild-type cells and either Gnas(E2-/E2-) or Gnas(+/E2-) cells. Gnas(E2-/E2-) chondrocytes phenocopied PTH/PTHrP receptor (PPR)(-/-) cells by prematurely undergoing hypertrophy. Introduction of a transgene expressing Gsalpha, one of several gene products that include Gnas exon 2, into Gnas(E2-/E2-) cells prevented premature hypertrophy. Gsalpha mRNA expression detected by real-time RT-PCR analysis was reduced to approximately half that of the wild-type in both paternal and maternal Gnas(+/E2-) growth plate chondrocytes, indicating biallelic expression of Gsalpha in these cells. Hypertrophy of Gnas(+/E2-) chondrocytes was modestly but significantly premature in chimeric growth plates of mice containing wild-type and Gnas(+/E2-) cells. These data suggest that Gsalpha is the primary mediator of the actions of PPR in growth plate chondrocytes and that there is haploinsufficiency of Gsalpha signaling in Gnas(+/E2-) chondrocytes.
Figures


Similar articles
-
Chondrocyte-specific knockout of the G protein G(s)alpha leads to epiphyseal and growth plate abnormalities and ectopic chondrocyte formation.J Bone Miner Res. 2005 Apr;20(4):663-71. doi: 10.1359/JBMR.041210. Epub 2004 Dec 6. J Bone Miner Res. 2005. PMID: 15765186
-
Shortened Fingers and Toes: GNAS Abnormalities are Not the Only Cause.Exp Clin Endocrinol Diabetes. 2020 Oct;128(10):681-686. doi: 10.1055/a-1047-0334. Epub 2019 Dec 11. Exp Clin Endocrinol Diabetes. 2020. PMID: 31860119 Free PMC article.
-
Coding GNAS mutations leading to hormone resistance impair in vitro agonist- and cholera toxin-induced adenosine cyclic 3',5'-monophosphate formation mediated by human XLalphas.Endocrinology. 2006 May;147(5):2253-62. doi: 10.1210/en.2005-1487. Epub 2006 Feb 16. Endocrinology. 2006. PMID: 16484323
-
[Cytokines in bone diseases. Genetic defects of PTH/PTHrP receptor in chondrodysplasia].Clin Calcium. 2010 Oct;20(10):1481-8. Clin Calcium. 2010. PMID: 20890029 Review. Japanese.
-
GNAS mutations and heterotopic ossification.Bone. 2018 Apr;109:80-85. doi: 10.1016/j.bone.2017.09.002. Epub 2017 Sep 6. Bone. 2018. PMID: 28889026 Free PMC article. Review.
Cited by
-
Pseudohypoparathyroidism Type 1A with Normocalcaemia, due to the Novel C.389A>G Variant of Exon 5 of the Guanine Nucleotide-Binding Protein, α-Stimulating Gene.J Bone Metab. 2021 Feb;28(1):85-89. doi: 10.11005/jbm.2021.28.1.85. Epub 2021 Feb 28. J Bone Metab. 2021. PMID: 33730787 Free PMC article.
-
Inhibitory function of parathyroid hormone-related protein on chondrocyte hypertrophy: the implication for articular cartilage repair.Arthritis Res Ther. 2012 Aug 31;14(4):221. doi: 10.1186/ar4025. Arthritis Res Ther. 2012. PMID: 22971952 Free PMC article. Review.
-
Coordination of chondrogenesis and osteogenesis by hypertrophic chondrocytes in endochondral bone development.J Bone Miner Metab. 2010 Sep;28(5):489-502. doi: 10.1007/s00774-010-0199-7. Epub 2010 Jul 6. J Bone Miner Metab. 2010. PMID: 20607327 Review.
-
Constitutive stimulatory G protein activity in limb mesenchyme impairs bone growth.Bone. 2018 May;110:230-237. doi: 10.1016/j.bone.2018.02.016. Epub 2018 Feb 20. Bone. 2018. PMID: 29471062 Free PMC article.
-
G-protein stimulatory subunit alpha and Gq/11α G-proteins are both required to maintain quiescent stem-like chondrocytes.Nat Commun. 2014 Apr 30;5:3673. doi: 10.1038/ncomms4673. Nat Commun. 2014. PMID: 24781502 Free PMC article.
References
-
- Jüppner, H., Abou-Samra, A. B., Freeman, M., Kong, X. F., Schipani, E., Richards, J., Kolakowski, L. F., Jr., Hock, J., Potts, J. T., Jr., Kronenberg, H. M., et al. (1991) Science 254, 1024–1026. - PubMed
-
- Karaplis, A. C., Luz, A., Glowacki, J., Bronson, R. T., Tybulewicz, V. L., Kronenberg, H. M. & Mulligan, R. C. (1994) Genes Dev. 8, 277–289. - PubMed
-
- Lanske, B., Karaplis, A. C., Lee, K., Luz, A., Vortkamp, A., Pirro, A., Karperien, M., Defize, L. H., Ho, C., Mulligan, R. C., et al. (1996) Science 273, 663–666. - PubMed
-
- Guo, J., Chung, U. I., Kondo, H., Bringhurst, F. R. & Kronenberg, H. M. (2002) Dev. Cell 3, 183–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

